library(tidyverse)
library(writexl)Solution 2: Summary Statistics and Data Wrangling
Introduction
In this exercise you will do some more advance tidyverse operations such as pivoting and nesting.
First steps
- Load packages.
- Load the joined diabetes data set you created in exercise 1 (e.g. “diabetes_join.xlsx”) and the glucose dataset
df_glucose.xlsxfrom the data folder. If you did not make it all the way through exercise 1 you can find the dataset you need indata/exercise1_diabetes_join.xlsx
diabetes_join <- readxl::read_excel('../data/exercise1_diabetes_join.xlsx')
df_glucose <- readxl::read_excel('../data/df_glucose.xlsx')Change format
- Have a look at the glucose dataset. It has three columns with measurements from a Oral Glucose Tolerance Test where blood glucose is measured at fasting (Glucose_0), 1 hour/60 mins after glucose intake (Glucose_6), and 2 hours/120 mins after (Glucose_120). The last columns is an ID column. Change the data type of the ID column to
factorin bothdiabetes_joinanddf_glucose.
df_glucose$ID <- as.factor(df_glucose$ID)
diabetes_join$ID <- as.factor(diabetes_join$ID)- Restructure the glucose dataset into a long format. Think about which columns should be included in the pivot.
df_glucose_long <- df_glucose %>%
pivot_longer(cols = starts_with("Glucose"),
names_to = "Measurement",
values_to = "Glucose (mmol/L)"
)
head(df_glucose_long)# A tibble: 6 × 3
ID Measurement `Glucose (mmol/L)`
<fct> <chr> <dbl>
1 9046 Glucose_0 6.65
2 9046 Glucose_60 8.04
3 9046 Glucose_120 10.0
4 51676 Glucose_0 4.49
5 51676 Glucose_60 5.40
6 51676 Glucose_120 6.22- How many rows are there per ID? Does that make sense?
# There are three rows for each ID, corresponding to the three glucose measurements
df_glucose_long %>%
count(ID) %>%
head()# A tibble: 6 × 2
ID n
<fct> <int>
1 129 3
2 210 3
3 491 3
4 530 3
5 621 3
6 712 3Change factor levels
- In your long formatted dataframe you should have one column that described which measurement the row refers to, i.e. Glucose_0, Glucose_60 or Glucose_120. Transform this column so you only have the numerical part, i.e. only 0, 60 or 120. Then change the data type of that column to
factor. Check the order of the factor levels and if necessary change them to the proper order.
Have a look at the help for factors ?factors to see how to influence the levels.
df_glucose_long <- df_glucose_long %>%
mutate(Measurement = str_split_i(Measurement, '_', 2) %>% as.factor())Check factor levels:
levels(df_glucose_long$Measurement)[1] "0" "120" "60" Adjust levels to proper order:
df_glucose_long$Measurement <- factor(df_glucose_long$Measurement, levels = c('0', '60','120'))Join datasets
- Join the long formatted glucose dataset you made in 4 with the joined diabetes dataset you loaded in 2. Do the wrangling needed to make it happen!
diabetes_glucose <- diabetes_join %>%
mutate(ID = str_split_i(ID, pattern = '_', i = 2)) %>%
left_join(df_glucose_long, by = 'ID')
head(diabetes_glucose)# A tibble: 6 × 13
ID Sex Age BloodPressure BMI PhysicalActivity Smoker Diabetes
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 34120 Female 28 75 25.4 92 Never 0
2 34120 Female 28 75 25.4 92 Never 0
3 34120 Female 28 75 25.4 92 Never 0
4 27458 Female 55 72 24.6 86 Never 0
5 27458 Female 55 72 24.6 86 Never 0
6 27458 Female 55 72 24.6 86 Never 0
# ℹ 5 more variables: Serum_ca2 <dbl>, Married <chr>, Work <chr>,
# Measurement <fct>, `Glucose (mmol/L)` <dbl>Summary stats
- Calculate the mean glucose levels for each time point.
You will need to use group_by() and summerise().
diabetes_glucose %>%
group_by(Measurement) %>%
summarise(mean = mean(`Glucose (mmol/L)`))# A tibble: 3 × 2
Measurement mean
<fct> <dbl>
1 0 8.07
2 60 9.71
3 120 11.0 - Make a figure with boxplots of the glucose measurements across the three time points.
diabetes_glucose %>%
ggplot(aes(y = `Glucose (mmol/L)`,
x = Measurement,
fill = Measurement)) +
geom_boxplot() +
theme_bw()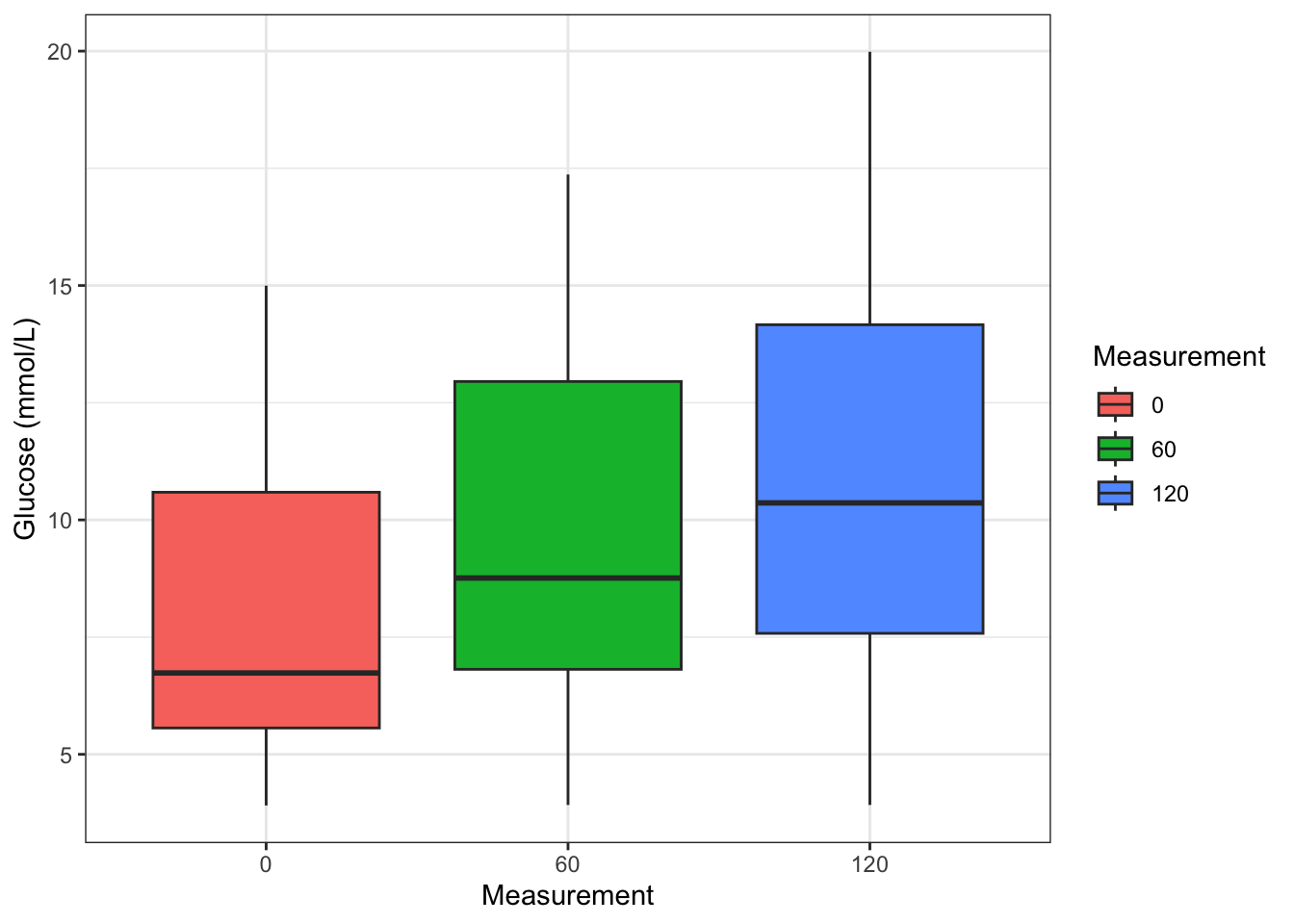
- Calculate mean and standard deviation for all numeric columns and reformat the data frame into a manageable format like we did in the presentation.
You will need to use summarise() and across() to select numeric columns.
For a nice reformatting like in the presentation with need both pivot_longer and pivot_wider.
diabetes_glucose %>%
summarise(across(.cols = where(is.numeric),
.fns = list(mean = ~ mean(., na.rm = TRUE),
sd = ~ sd(., na.rm = TRUE)),
.names = "{.col}-{.fn}")) %>%
pivot_longer(cols = everything(),
names_to = c("variable", "statistic"),
names_sep = "-") %>%
pivot_wider(names_from = statistic,
values_from = value)# A tibble: 7 × 3
variable mean sd
<chr> <dbl> <dbl>
1 Age 33.8 11.4
2 BloodPressure 72.6 13.0
3 BMI 30.1 6.67
4 PhysicalActivity 84.1 25.3
5 Diabetes 0.502 0.500
6 Serum_ca2 9.44 0.257
7 Glucose (mmol/L) 9.60 3.76 - Plot
PhysicalActivityandBMIagainst each other in a scatter plot and color byGlucose (mmol/L).
diabetes_glucose %>%
ggplot(aes(x = PhysicalActivity,
y = BMI,
color = `Glucose (mmol/L)`)) +
geom_point() +
theme_bw()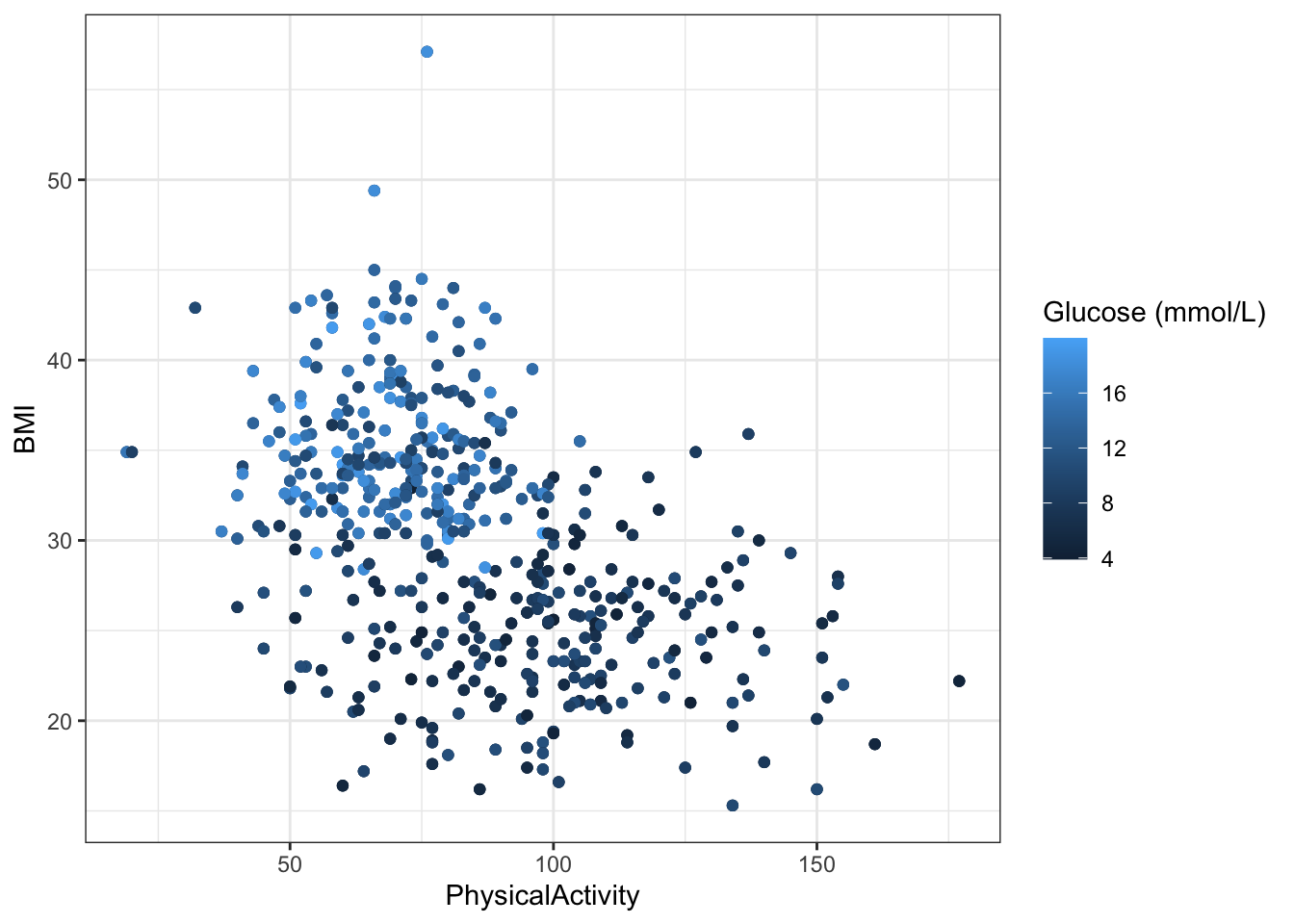
- Highlights any trends or patterns that emerge across
PhysicalActivityandBMIvalues. Includes an indication of confidence around those trends. Change color palette.
diabetes_glucose %>%
ggplot(aes(x = PhysicalActivity,
y = BMI,
color = `Glucose (mmol/L)`)) +
geom_point() +
geom_smooth(method = "loess", level = 0.95) +
labs(title = "Glucose Measurements with Mean by Diabetes Status") +
scale_color_viridis_c(option = "C") +
theme_bw()`geom_smooth()` using formula = 'y ~ x'Warning: The following aesthetics were dropped during statistical transformation:
colour.
ℹ This can happen when ggplot fails to infer the correct grouping structure in
the data.
ℹ Did you forget to specify a `group` aesthetic or to convert a numerical
variable into a factor?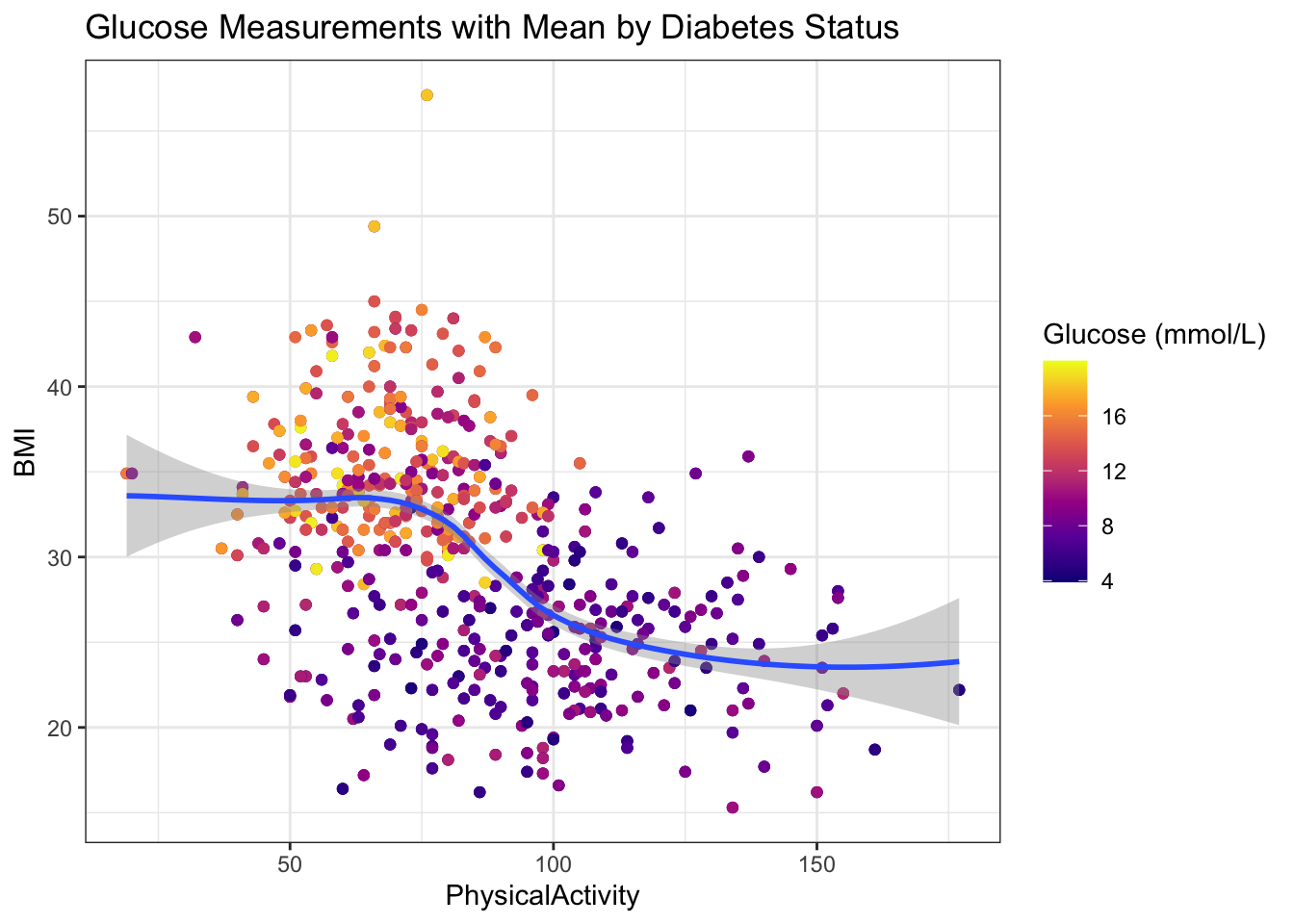
Sample outliers across variables
- Just like we did in the presentation, make a histogram for each numeric variable in the dataset. You can copy-paste the code from the presentation and make very few changes to make it work in this case.
Check the class of the variables in the dataset.
diabetes_glucose_num <- diabetes_glucose %>%
select(where(is.numeric)) %>%
drop_na()
diabetes_glucose_num[1:5, ]# A tibble: 5 × 7
Age BloodPressure BMI PhysicalActivity Diabetes Serum_ca2
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 28 75 25.4 92 0 9.6
2 28 75 25.4 92 0 9.6
3 28 75 25.4 92 0 9.6
4 55 72 24.6 86 0 9.2
5 55 72 24.6 86 0 9.2
# ℹ 1 more variable: `Glucose (mmol/L)` <dbl>diabetes_glucose_num %>%
pivot_longer(cols = where(is.numeric),
names_to = "variable",
values_to = "value") %>%
ggplot(., aes(x = value)) +
geom_histogram(color= 'white') +
facet_wrap(vars(variable), ncol = 2, scales = "free") +
theme_minimal()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.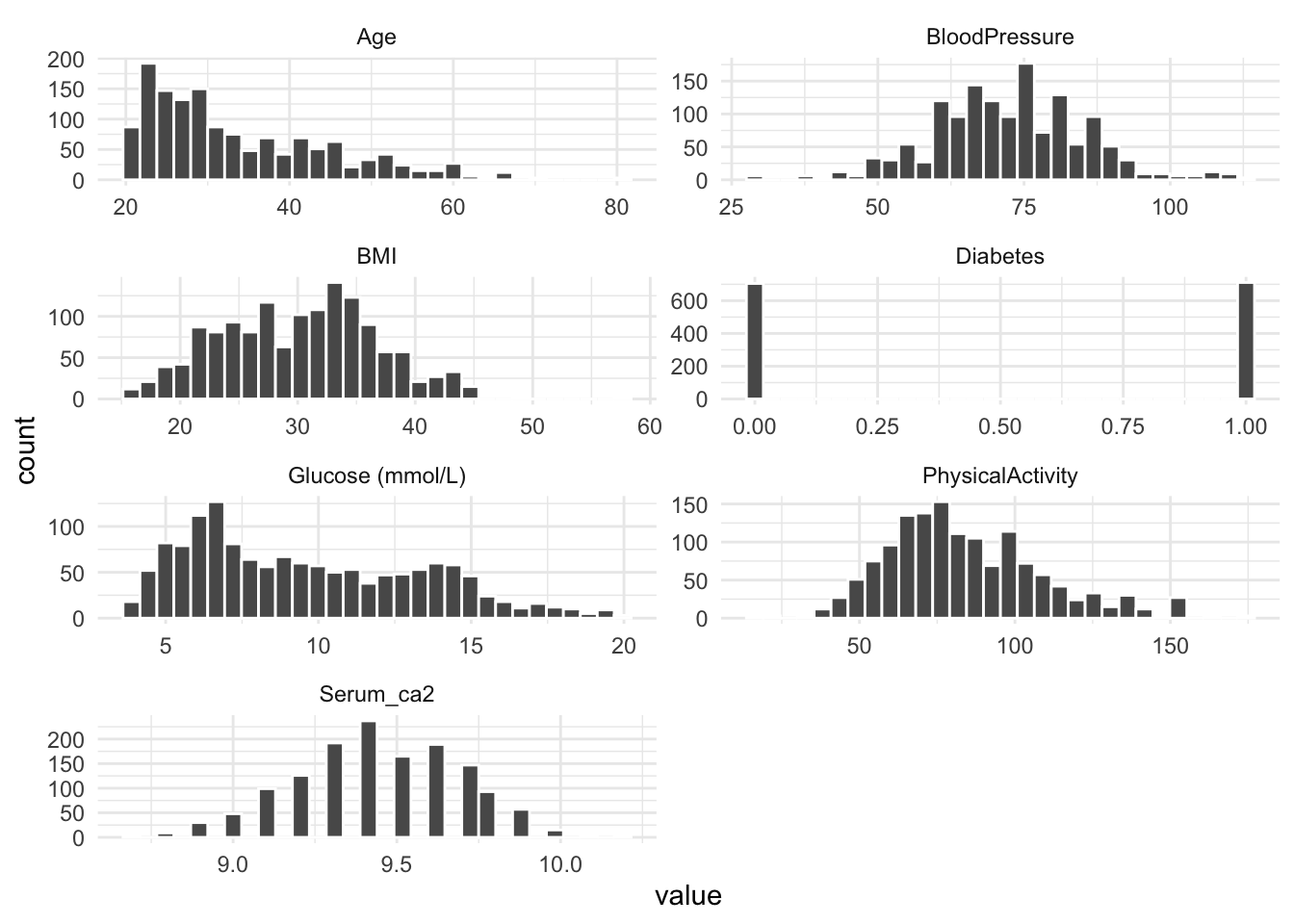
- Check for outlier samples using the dendogram showed in the presentation. This includes scaling, calculating the distances between all pairs of scaled variables and hierarchical clustering. Remember that this can only be done for the numeric variables.
We will not spot any outliers via the dendogram in this dataset so we will add a fake outlier for you to detect. Here, the outlier has a value of the maximum value + 5 in each of the variables - pretty unrealistic. Play around with the values of the fake outlier and see how extreme it needs to be to be spotted in the dendogram.
outlier <- tibble(Age = max(diabetes_glucose_num$Age) + 5,
BloodPressure = max(diabetes_glucose_num$BloodPressure) + 5,
BMI = max(diabetes_glucose_num$BMI) + 5,
PhysicalActivity = max(diabetes_glucose_num$PhysicalActivity) + 5,
Serum_ca2 = max(diabetes_glucose_num$Serum_ca2) + 5,
`Glucose (mmol/L)` = max(diabetes_glucose_num$`Glucose (mmol/L)`) + 5,
)
# Add a new row
diabetes_glucose_num_outlier <- add_row(diabetes_glucose_num, outlier)
# Keep adding outliters
# diabetes_glucose_num_outlier <- add_row(diabetes_glucose_num_outlier, outlier)# Euclidean pairwise distances
diabetes_glucose_dist <- diabetes_glucose_num_outlier %>%
mutate(across(everything(), scale)) %>%
dist(., method = 'euclidean')
# Hierarchical clustering with Ward's distance metric
hclust_num <- hclust(diabetes_glucose_dist, method = 'ward.D2')
plot(hclust_num, main="Clustering based on scaled integer values", cex=0.7)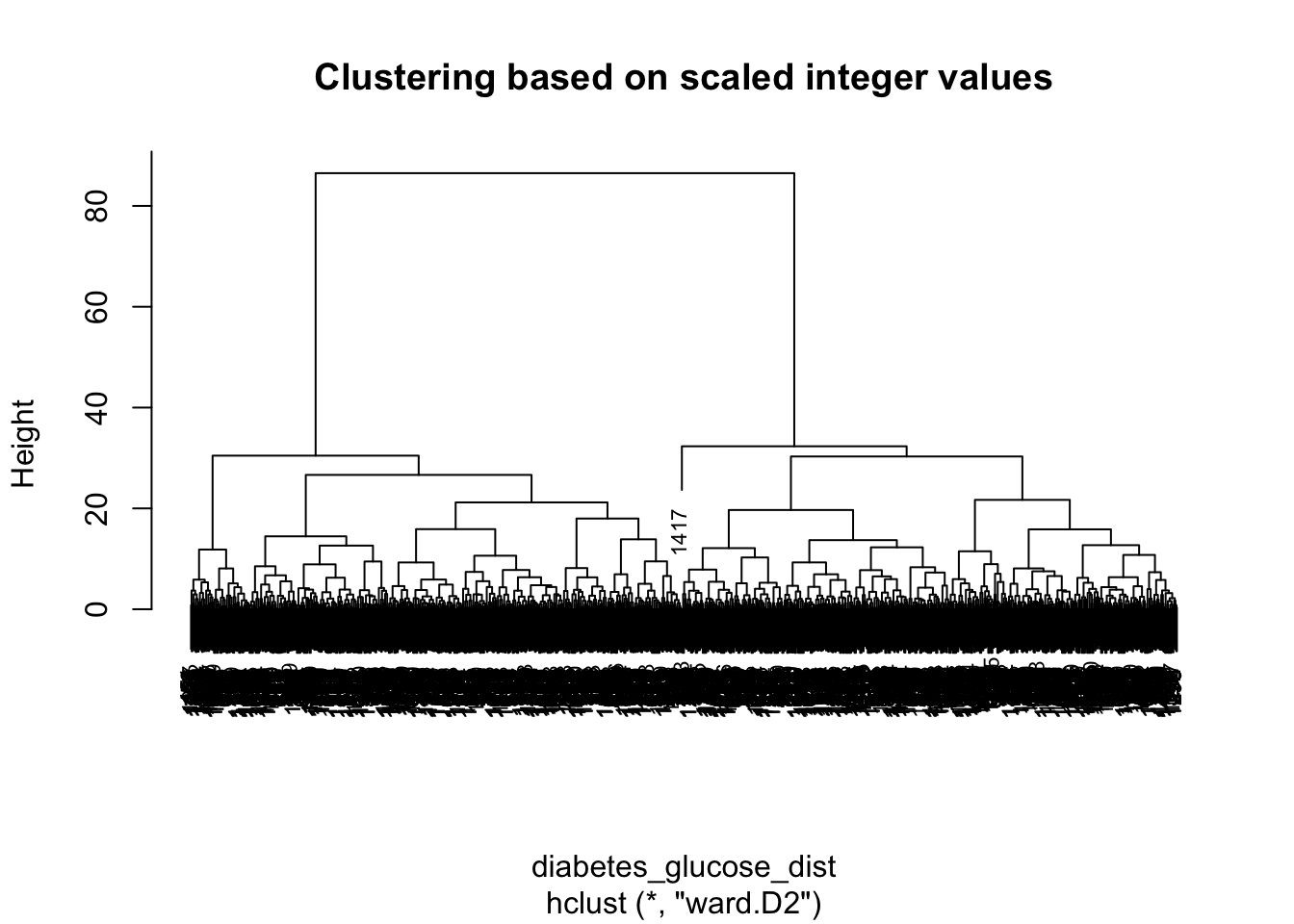
- Export the final dataset (without the outlier!) in whichever format you prefer.
writexl::write_xlsx(diabetes_glucose, '../data/exercise2_diabetes_glucose.xlsx')More Plotting
Let’s make some more advanced ggplots!
- For p1, plot the density of the glucose measurements at time 0. For p2, make the same plot as p1 but add stratification on the diabetes status. Make sure Diabetes is a factor. Give the plot a meaningful title. Consider the densities - do the plots make sense?
diabetes_glucose$Diabetes <- as.factor(diabetes_glucose$Diabetes)
p1 <- diabetes_glucose %>%
filter(Measurement == 0) %>%
ggplot(aes(x = `Glucose (mmol/L)`)) +
geom_density()
p2 <- diabetes_glucose %>%
filter(Measurement == 0) %>%
ggplot(aes(x = `Glucose (mmol/L)`,
linetype = Diabetes)) +
geom_density()
library(patchwork)
p1 + p2 + plot_annotation(title = "Glucose measurements at time 0")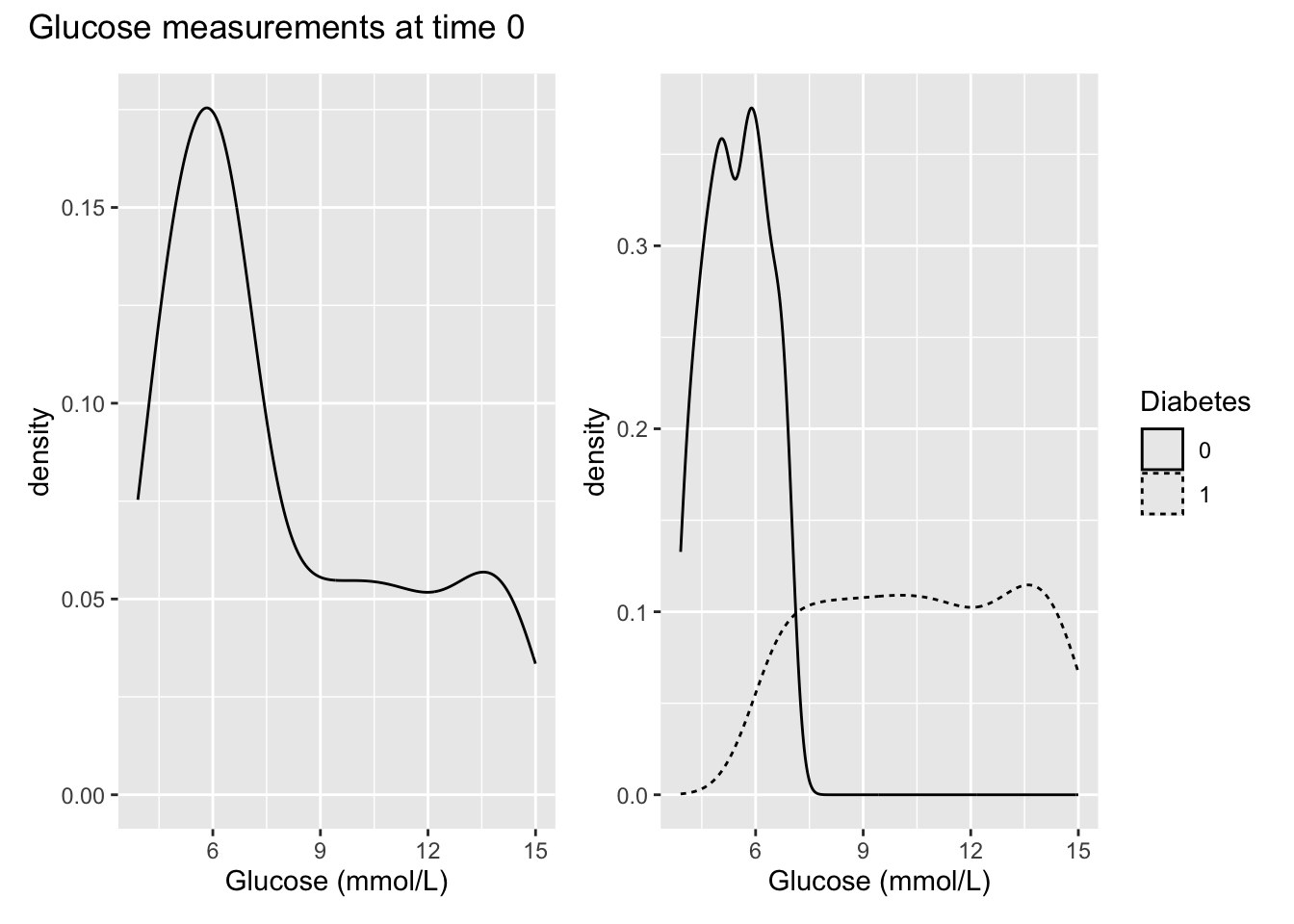
- Now, create one plot for the glucose measurement where the densities are stratified on measurement time (0, 60, 120) and diabetes status (0, 1). You should see 6 density curves in your plot. ::: {.callout-tip collapse=“true”} ## Hint There two ways of stratifying a density plot in ggplot2:
colorandlinetype. :::
p1 <- diabetes_glucose %>%
ggplot(aes(x = `Glucose (mmol/L)`,
color = Measurement,
linetype = Diabetes)) +
geom_density() +
labs(title = "Glucose measurements at time 0, 60, and 120 across diabetes status")
p1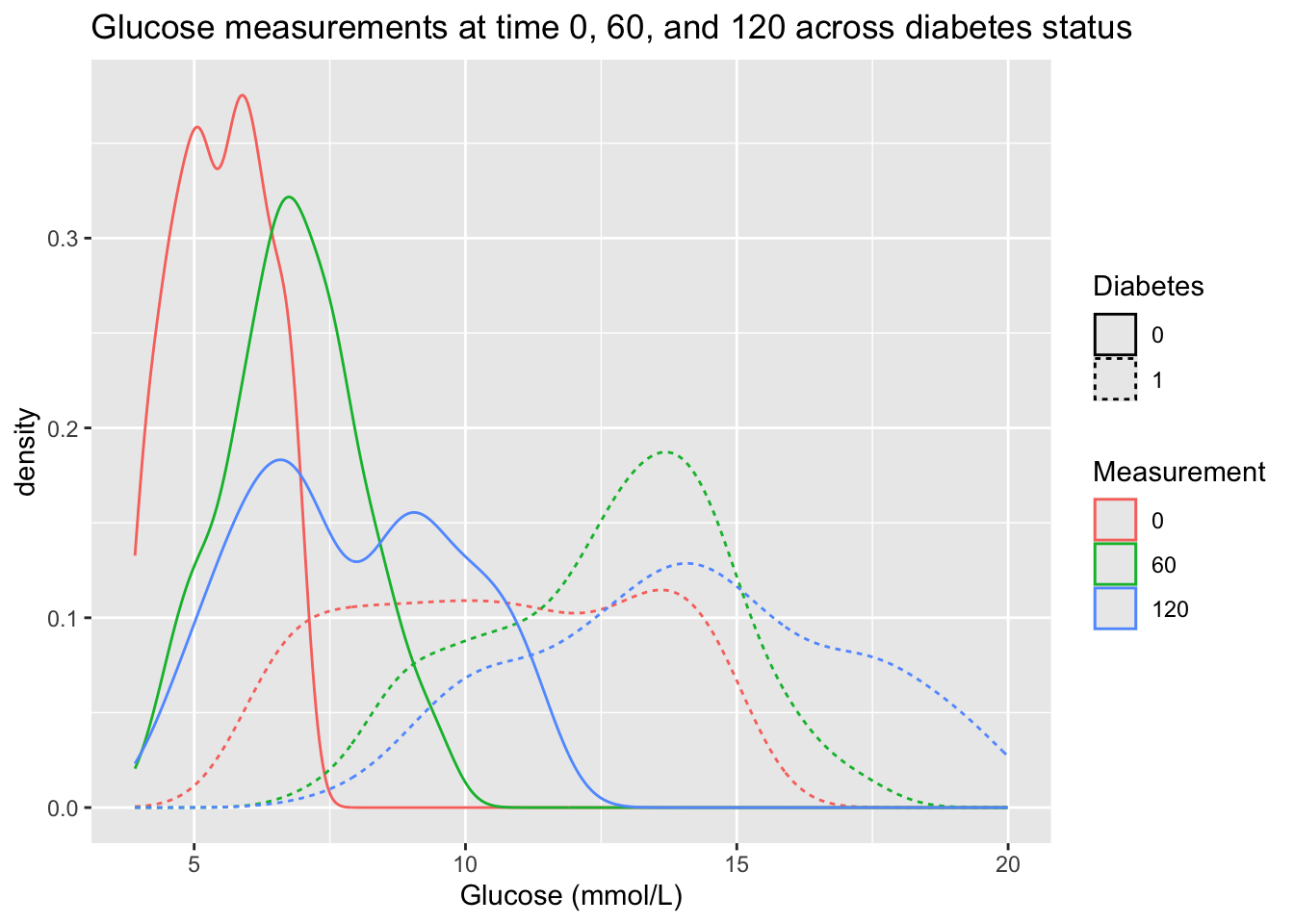
- That is not a very intuitive plot. Could we change the geom and the order of the variables to visualize the same information more intuitively?
p1 <- diabetes_glucose %>%
ggplot(aes(x = Diabetes, y = `Glucose (mmol/L)`,
color = Diabetes,
linetype = Measurement)) +
geom_boxplot() +
labs(title = "Glucose measurements at time 0, 60, and 120 across diabetes status")
p1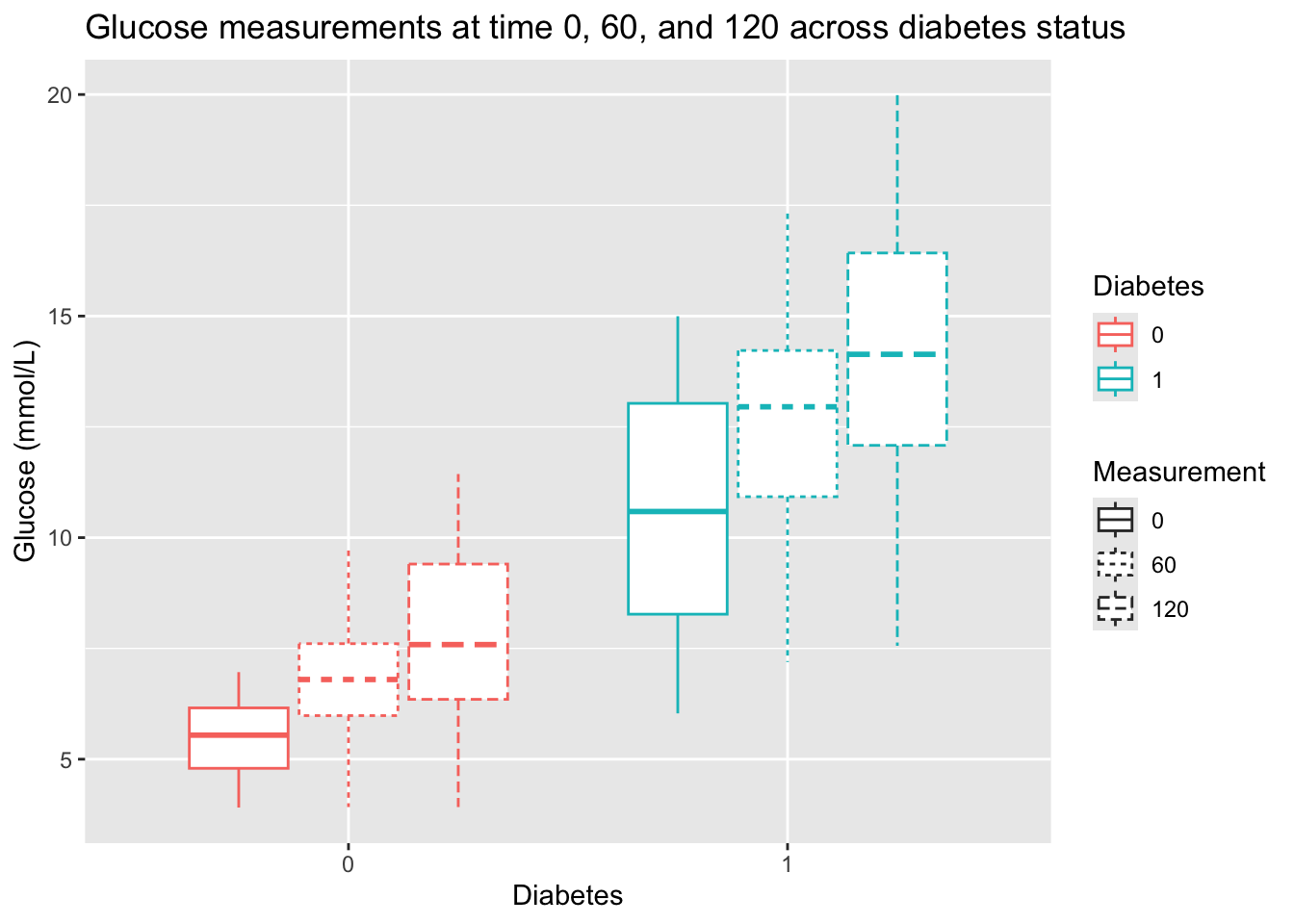
- Calculate mean Glucose level by time point and
Diabetesstatus. Save the data frame in a variable.
Group by several variables: group_by(var1, var2).
glucose_mean <- diabetes_glucose %>%
group_by(Measurement, Diabetes) %>%
summarize(`Glucose (mmol/L)` = mean(`Glucose (mmol/L)`)) %>%
ungroup()`summarise()` has grouped output by 'Measurement'. You can override using the
`.groups` argument.glucose_mean# A tibble: 6 × 3
Measurement Diabetes `Glucose (mmol/L)`
<fct> <fct> <dbl>
1 0 0 5.49
2 0 1 10.6
3 60 0 6.82
4 60 1 12.6
5 120 0 7.88
6 120 1 14.2 - Create a plot that visualizes glucose measurements across time points, with one line for each patient ID. Then color the lines by their diabetes status. In summary, each patient’s glucose measurements should be connected with a line, grouped by their ID, and color-coded by
Diabetes. Give the plot a meaningful title.
diabetes_glucose %>%
ggplot(aes(x = Measurement,
y = `Glucose (mmol/L)`)) +
geom_point(aes(color = Diabetes)) +
geom_line(aes(group = ID, color = Diabetes)) +
labs(title = 'Glucose Measurements Across Time Points by Diabetes Status')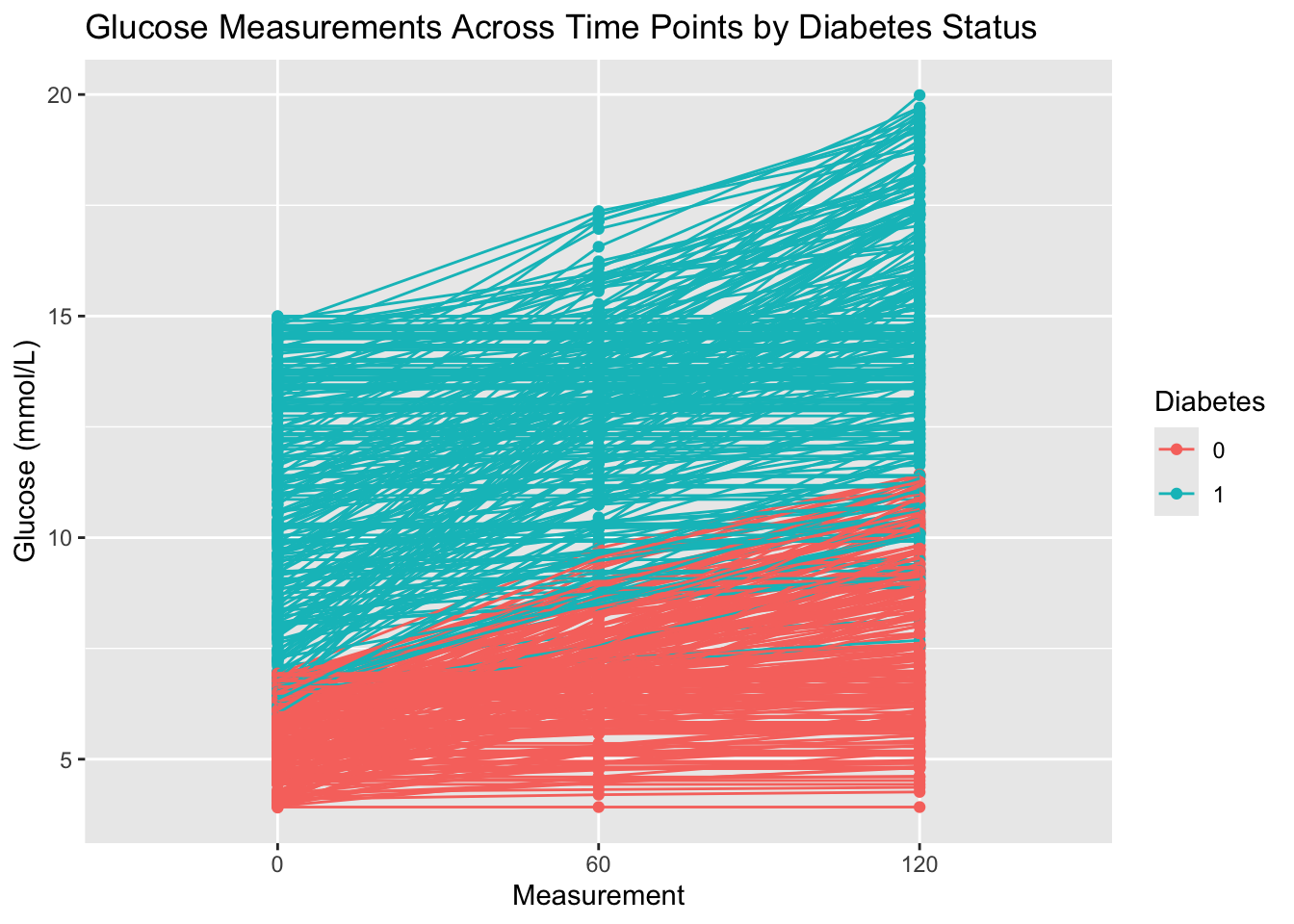
- Recreate the plot you made above.
But this time: Jittered individual values to avoid over plotting. Overlay and connect the group means showing the mean trend for each group. Add a title, change color pallet and themes.
This plot should look like this:
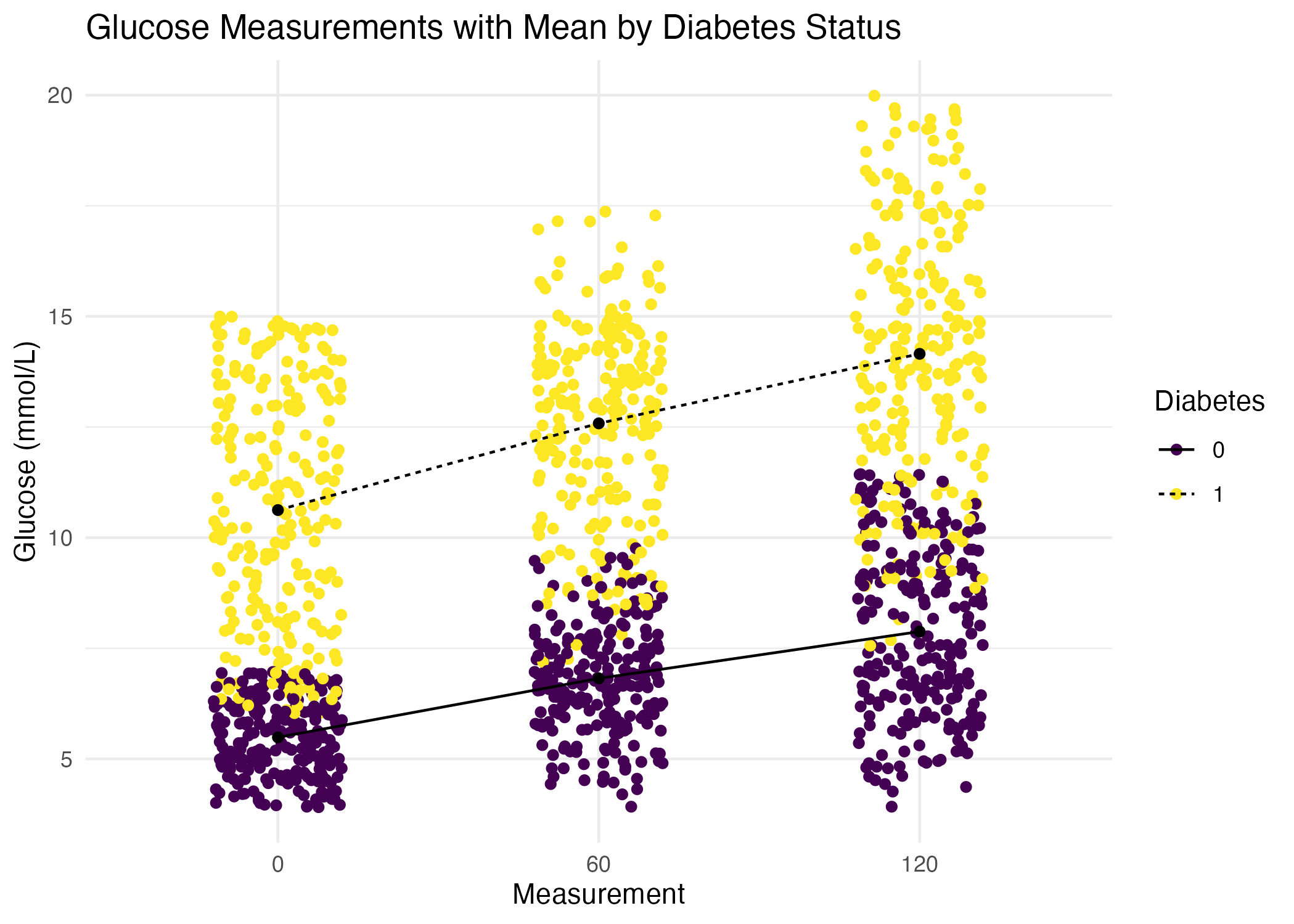
diabetes_glucose %>%
ggplot(aes(x = Measurement,
y = `Glucose (mmol/L)`)) +
geom_jitter(aes(color = Diabetes), width = 0.2) +
geom_point(data = glucose_mean, aes(x = Measurement, y = `Glucose (mmol/L)`)) +
geom_line(data = glucose_mean, aes(x = Measurement, y = `Glucose (mmol/L)`,
group = Diabetes, linetype = Diabetes)) +
labs(title = "Glucose Measurements with Mean by Diabetes Status") +
scale_colour_manual(values = c("#440154FF", "#FDE725FF")) +
theme_minimal() 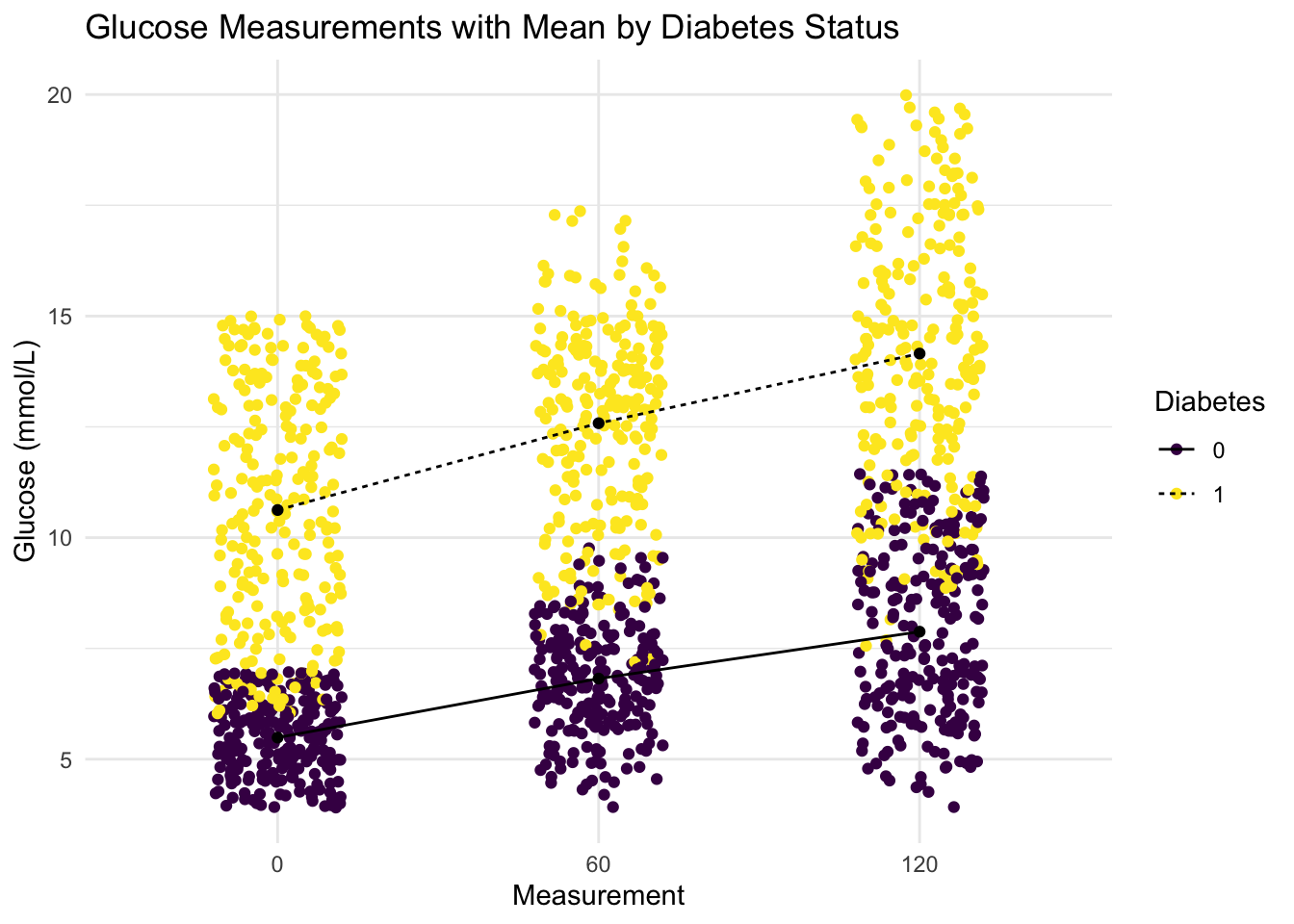
ggsave('../figures/figure3_13.png')Saving 7 x 5 in image