library(tidyverse)
library(factoextra)Presentation 2: Summary Statistics and Data Wrangling
Now that we’ve cleaned our data, it’s time to dig deeper into the actual contents of our dataset. Lets do some Exploratory Data Analysis (EDA).
Load Packages and Data
Let’s begin by loading the packages we’ll need for data wrangling and plotting:
Next, bring in the cleaned dataset we prepared earlier:
load("../data/Ovarian_comb_clean.RData")
# Check the structure and first few rows
class(df_comb)[1] "tbl_df" "tbl" "data.frame"df_comb %>% slice(1:5)# A tibble: 5 × 54
unique_patient_ID sample_recode primarysite summarygrade tumorstage substage
<chr> <chr> <fct> <fct> <fct> <chr>
1 TCGA-20-0987 Tumor ov high 3 c
2 TCGA-23-1031 Tumor ov high 4 <NA>
3 TCGA-24-0979 Tumor ov high 4 <NA>
4 TCGA-23-1117 Tumor ov high 3 c
5 TCGA-23-1021 Tumor ov high 4 <NA>
# ℹ 48 more variables: grade <fct>, age_at_initial_path_diagn <int>,
# days_to_tumor_recurrence <int>, recurrence_status <fct>,
# days_to_death <int>, vital_status <fct>, percent_normal_cells <int>,
# percent_stromal_cells <int>, percent_tumor_cells <int>, batch <fct>,
# percent_not_cancer_cells <int>, stage <chr>, dominant_cell_type <chr>,
# group <chr>, COL10A1 <dbl>, COL11A1 <dbl>, COL11A2 <dbl>, COL13A1 <dbl>,
# COL14A1 <dbl>, COL15A1 <dbl>, COL16A1 <dbl>, COL17A1 <dbl>, …df_comb <- df_comb %>%
select(-percent_not_cancer_cells)Data Overview and ggplot2 Recap
Depending on how we want to investigate the variables, there are many ways in EDA toolkit to explore variables:
Use histograms or boxplots for single variables
Use scatterplots or barplots to check relationships between variables
Let’s revisit some of the variables. This is also a good chance to refresh your ggplot2 skills. If you need a detailed refresher, refer to the From Excel to R: Presentation 3.
# Distribution of tumor cell percentage
ggplot(df_comb,
aes(x = percent_tumor_cells)) +
geom_histogram(bins = 30, fill="#482878FF") +
theme_bw()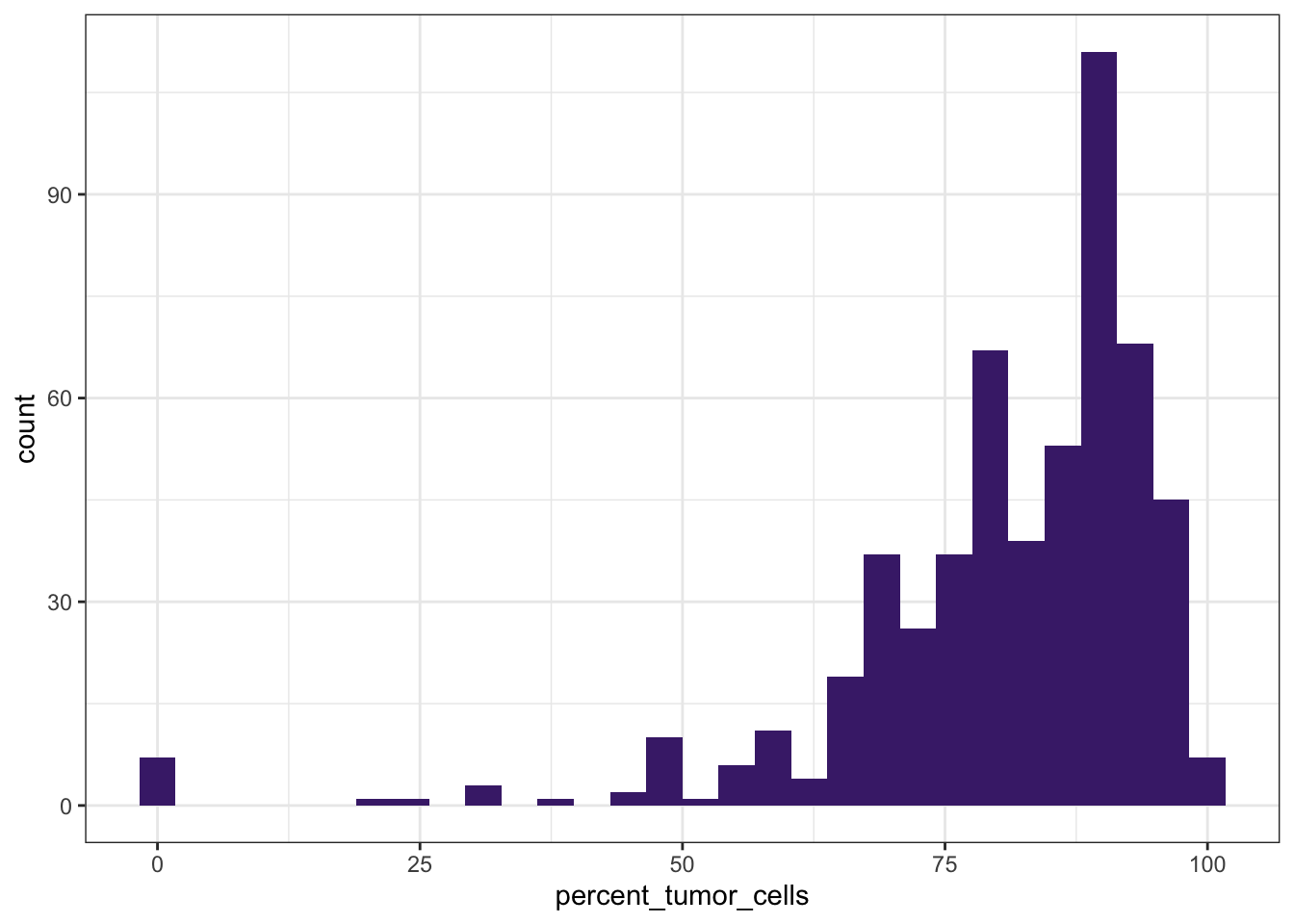
# Count with barplot
ggplot(df_comb,
aes(x = grade,
fill = grade)) +
geom_bar() +
theme_bw() +
scale_fill_viridis_d(na.value = "gray90")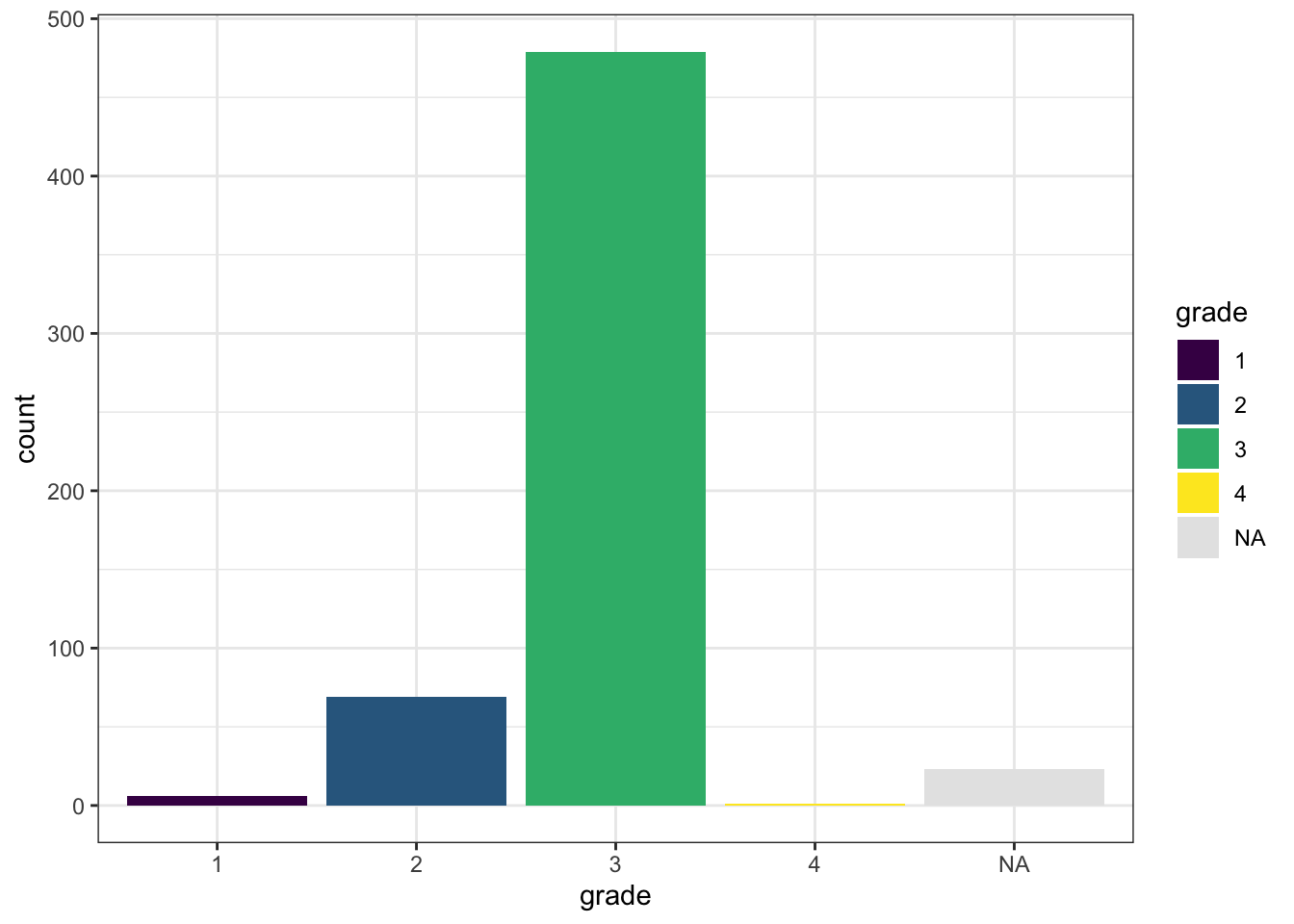
# Tumor percentage by summary grade
ggplot(df_comb,
aes(x = grade,
y = percent_tumor_cells,
fill = grade)) +
geom_boxplot() +
theme_bw() +
scale_fill_viridis_d(na.value = "gray90")Warning: Removed 22 rows containing non-finite outside the scale range
(`stat_boxplot()`).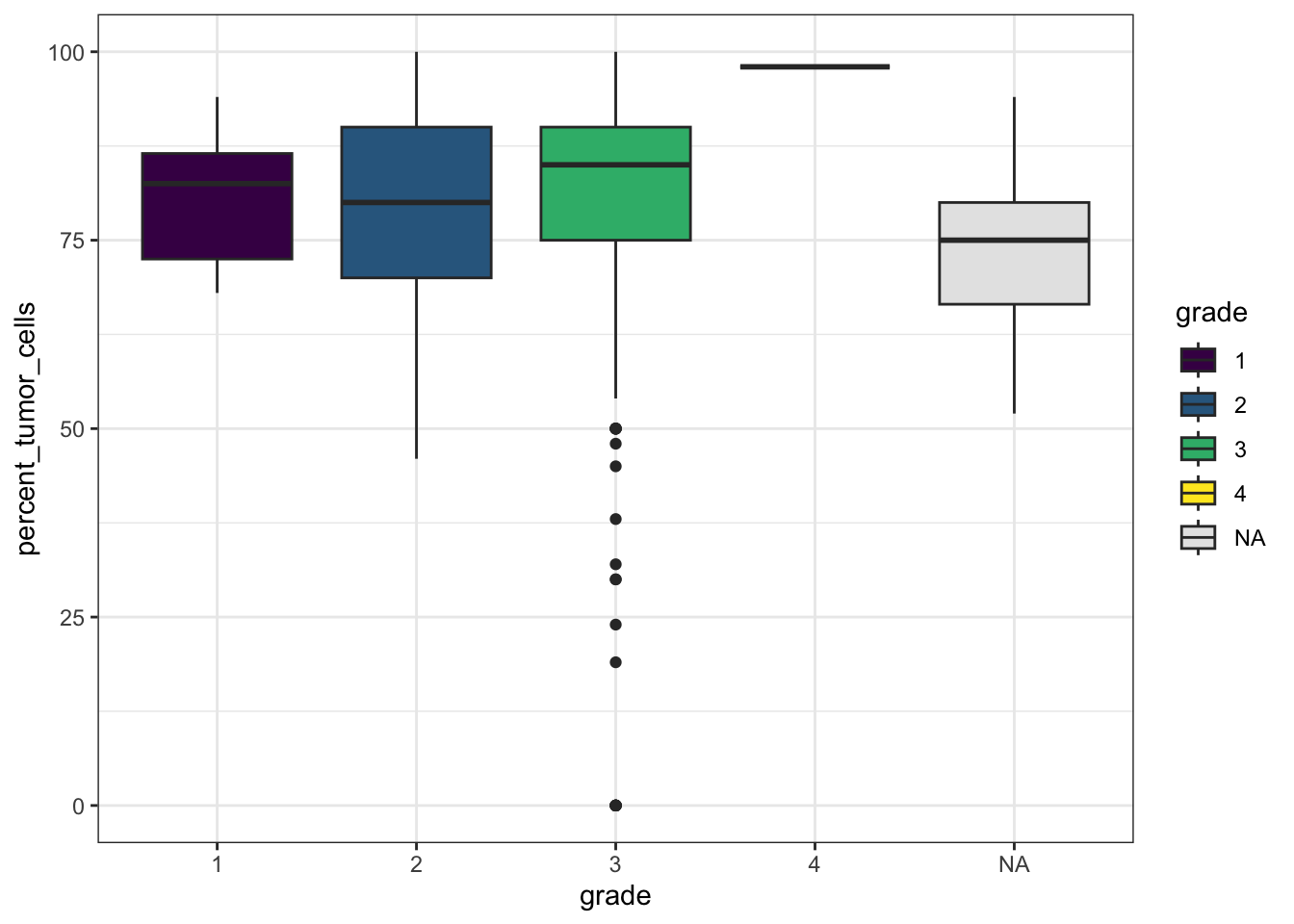
These simple plots give us an initial overview of the distribution of some of the variables - on their own and stratified by groups.
Formats: long and wide
Now, let’s say we want to create a single plot and compare several COL gene expression levels across different categories.
To plot and analyze more effectively, we need to reshape the data into long format, where each row represents a single observation:
The data can be reformatted to long format using pivot_longer():
df_comb_long <- df_comb %>%
pivot_longer(cols = starts_with('COL1'),
names_to = "gene",
values_to = "value")
df_comb_long %>% select(unique_patient_ID, age_at_initial_path_diagn, gene, value) %>% head() # A tibble: 6 × 4
unique_patient_ID age_at_initial_path_diagn gene value
<chr> <int> <chr> <dbl>
1 TCGA-20-0987 61 COL10A1 3.66
2 TCGA-20-0987 61 COL11A1 3.20
3 TCGA-20-0987 61 COL11A2 3.65
4 TCGA-20-0987 61 COL13A1 4.07
5 TCGA-20-0987 61 COL14A1 4.05
6 TCGA-20-0987 61 COL15A1 4.98# one line per gene per person
nrow(df_comb)[1] 578nrow(df_comb_long)[1] 6936The Long Format is ggplot’s Best Friend
With the reshaped df_comb_long, we can now create one combined plot that shows distributions for all genes in a single ggplot call. More context: add color-stratification by summarygrade and compare distributions side-by-side:
ggplot(na.omit(df_comb_long),
aes(x = gene, y = value, fill = summarygrade)) +
geom_boxplot() +
scale_fill_viridis_d() +
theme_minimal() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))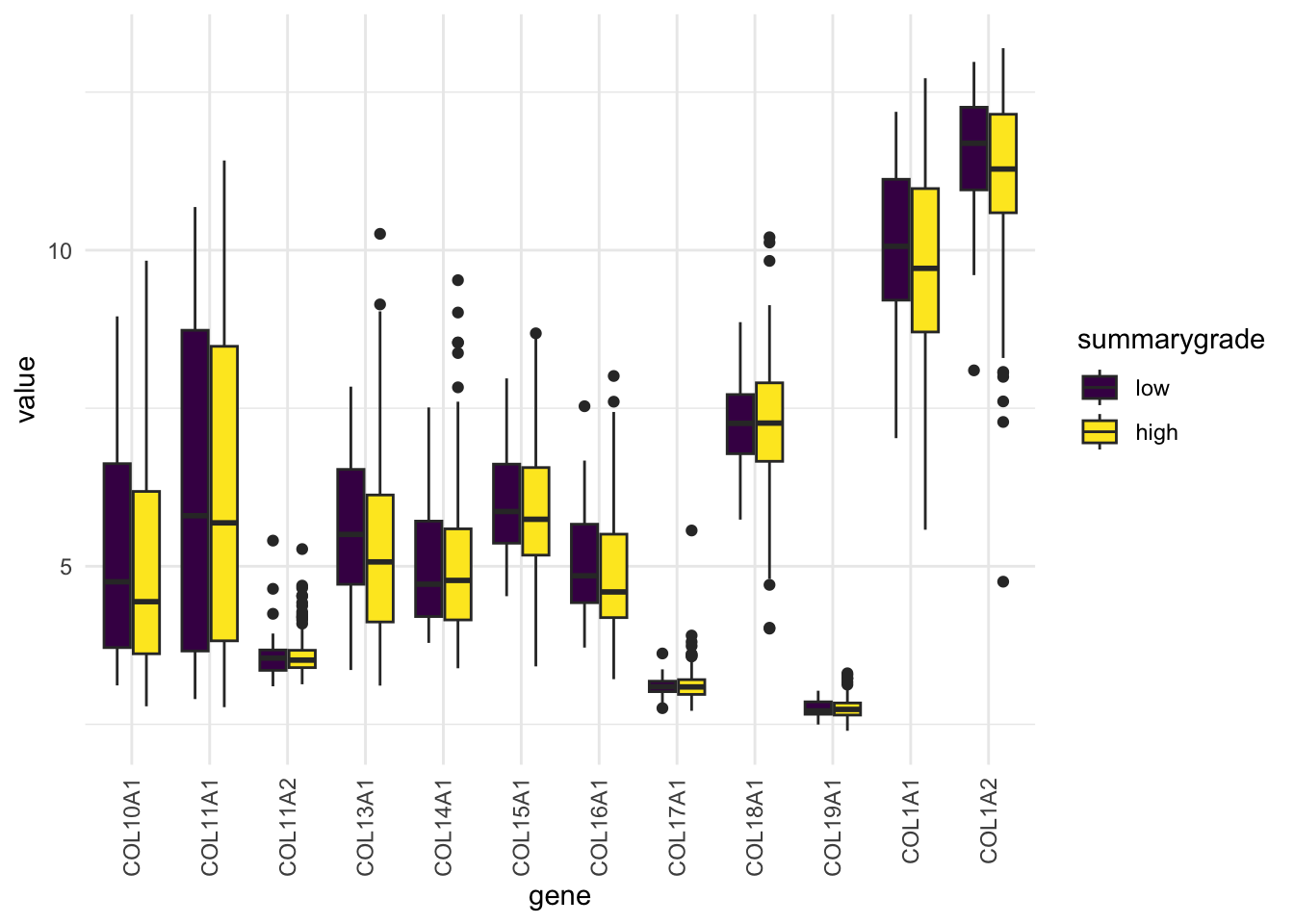
Want histograms for all genes?
ggplot(df_comb_long,
aes(x = value, fill = gene)) +
geom_histogram(bins = 30) +
facet_wrap(vars(gene), nrow = 3) +
scale_fill_viridis_d() +
theme_minimal()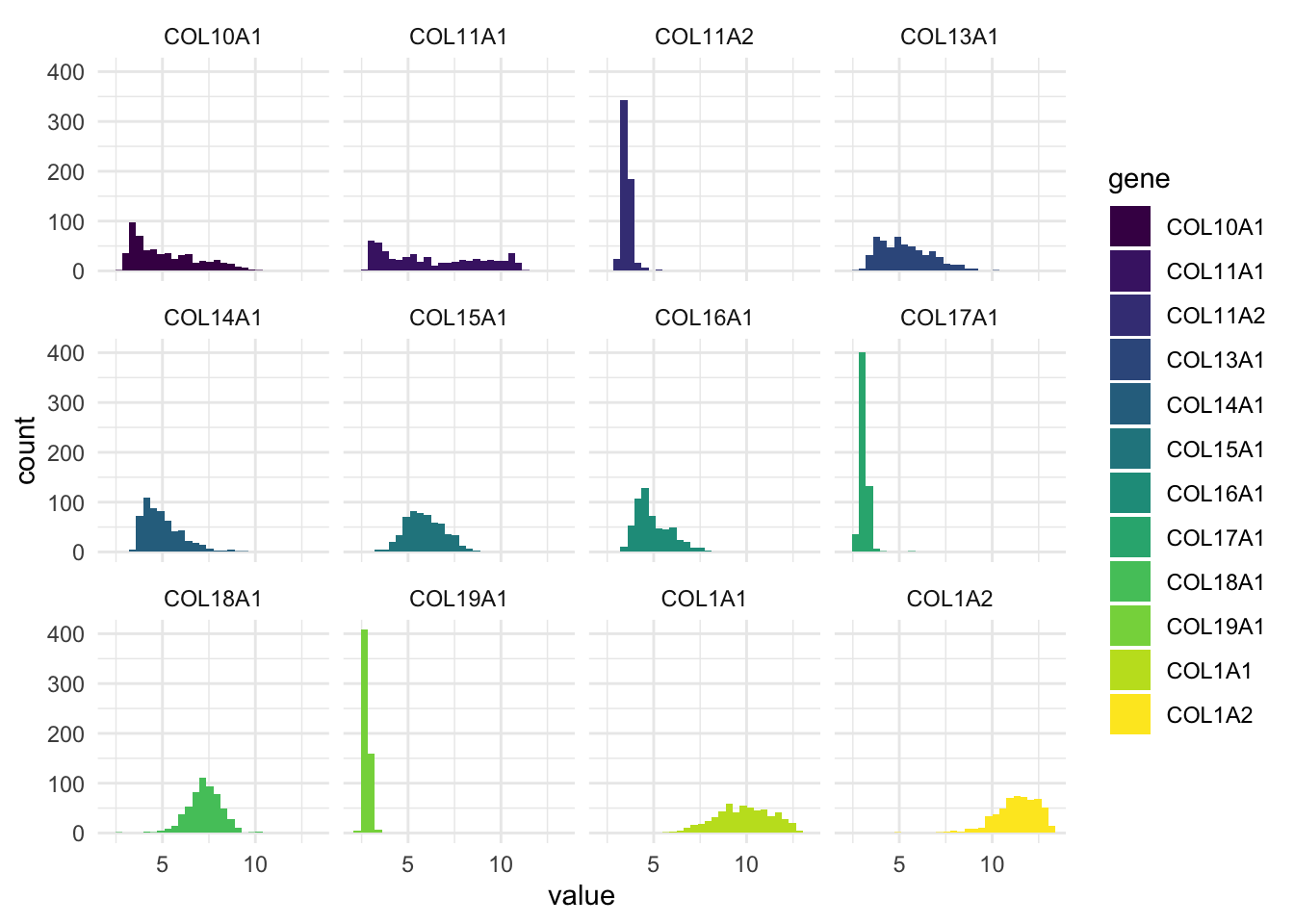
This plot gives us a histogram for each gene, all in one go.
No need to write separate plots manually.
Much easier to compare variables side-by-side.
Pivot back into wide format
The pivot_wider function is used to transform data to wide format.
df_comb_wide <- df_comb_long %>%
pivot_wider(names_from = gene,
values_from = value)
head(df_comb_wide)
head(df_comb)Exploring Factors (Categorical Variables)
When working with factor variables, we often want to know:
Which levels (categories) exist
Whether groups are balanced
How missing data overlaps
Let’s have a look at the vital_status variable. It looks fairly balanced.
table(df_comb$vital_status, useNA = "ifany")
deceased living <NA>
290 270 18 Now, let’s see how the vital_status levels are distributed across the tumorstage levels.
table(df_comb$vital_status, df_comb$stage, useNA = "ifany")
1-b 1-c 2-b 2-c 3-b 3-c 4-NA NA-NA
deceased 1 2 2 5 12 214 52 2
living 2 11 2 18 12 191 32 2
<NA> 0 0 0 0 0 7 0 11Some stage categories could be merged for more power and clearer groups:
# Relabel factor levels
df_comb <- df_comb %>%
mutate(stage = fct_recode(stage,
"1" = "1-b",
"1" = "1-c",
"2" = "2-b",
"2" = "2-c",
"4" = "4-NA",
NULL = "NA-NA"))Let’s see how other categorical variables relate to vital_status:
# Plot faceted bar plots colored by vital_status
df_comb %>%
select(where(is.factor), vital_status) %>%
drop_na() %>%
pivot_longer(cols = -vital_status, names_to = "variable", values_to = "value") %>%
ggplot(aes(x = value, fill = vital_status)) +
geom_bar() +
facet_wrap(vars(variable), scales = "free_x") +
scale_fill_viridis_d() +
theme_minimal() 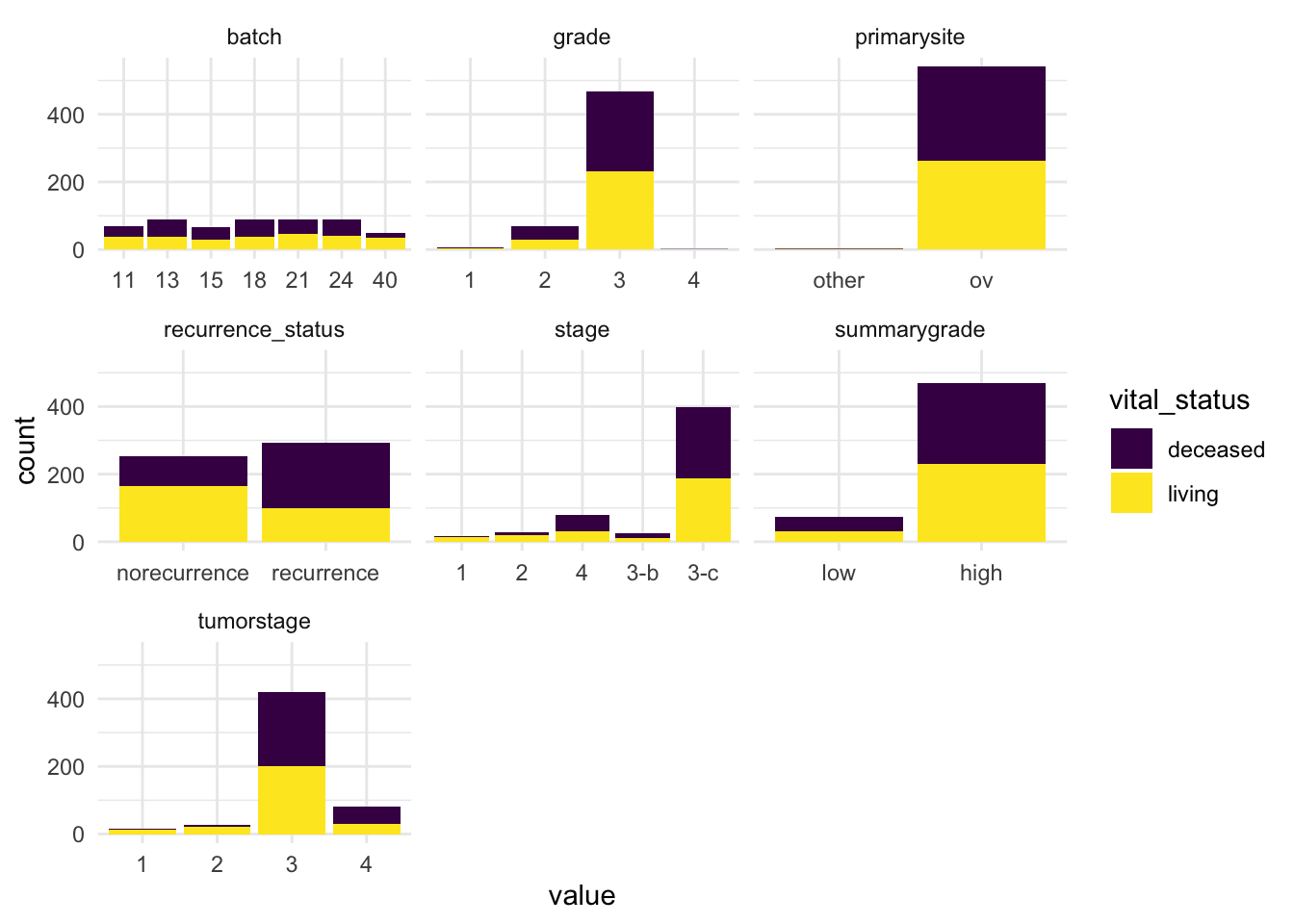
After reviewing our categorical variables, we observed the following:
Tumorstageandgradeare unbalanced. With certain stages and grades (e.g., 1 or 4) having low sample counts. This limits our ability to make reliable comparisons across all detailed levels.Batchis evenly represented, which reduces concerns about batch effects.Vital_statusis nicely balanced making it well-suited for comparisons and modeling.
These insights help us design our downstream analysis in a statistically sound and interpretable way.
Summary Statistics
It’s important to make sure our variables are well-behaved and ready for downstream analysis and modeling.
For this, let’s say we want to compute the mean of several columns. A basic (but tedious) approach might look like this:
df_comb %>%
summarise(mean_COL10A1 = mean(COL10A1),
mean_COL11A1 = mean(COL11A1),
mean_COL13A1 = mean(COL13A1),
mean_COL17A1 = mean(COL17A1),
mean_age_at_diagn = mean(age_at_initial_path_diagn))# A tibble: 1 × 5
mean_COL10A1 mean_COL11A1 mean_COL13A1 mean_COL17A1 mean_age_at_diagn
<dbl> <dbl> <dbl> <dbl> <dbl>
1 5.20 6.38 5.34 3.10 NAThis works, but we need to name every column we want to apply summarise to. It’s verbose and error-prone — especially if you have dozens of variables.
Instead of looking at individual variable, we’ll introduce some tidyverse helper functions: across(), where()and starts_with() - that make summarizing variables much more efficient and scalable — so you don’t have to write repetitive code for every column.
These helpers are useful when we want to apply a functions, i.e. summarise() or mutate() to several columns.
Using across() and everything() to select columns
Lets select the columns which we want to apply summarise across in a dynamic fashion:
df_comb %>%
summarise(across(.cols = everything(), # Columns to run fuction on
.fns = mean)) %>% # Function
select(15:25)# A tibble: 1 × 11
percent_tumor_cells batch stage dominant_cell_type group COL10A1 COL11A1
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 NA NA NA NA NA 5.20 6.38
# ℹ 4 more variables: COL11A2 <dbl>, COL13A1 <dbl>, COL14A1 <dbl>,
# COL15A1 <dbl>Using where() and starts_with() to select numeric columns
We will probably not want to calculate means on non-numeric columns, so let’s select only numeric columns. For that we need another helper caller where() that lets us select columns based on their properties, like data type.
df_comb %>%
summarise(across(.cols = where(fn = is.numeric),
.fns = mean)) %>%
select(5:15)# A tibble: 1 × 11
percent_stromal_cells percent_tumor_cells COL10A1 COL11A1 COL11A2 COL13A1
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 NA NA 5.20 6.38 3.55 5.34
# ℹ 5 more variables: COL14A1 <dbl>, COL15A1 <dbl>, COL16A1 <dbl>,
# COL17A1 <dbl>, COL18A1 <dbl>We can use starts_with() to select only columns starting with ‘COL’:
# Columns that start with "COL"
df_comb %>%
summarise(across(starts_with('COL'), mean))# A tibble: 1 × 34
COL10A1 COL11A1 COL11A2 COL13A1 COL14A1 COL15A1 COL16A1 COL17A1 COL18A1
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 5.20 6.38 3.55 5.34 5.01 5.89 4.95 3.10 7.26
# ℹ 25 more variables: COL19A1 <dbl>, COL1A1 <dbl>, COL1A2 <dbl>,
# COL21A1 <dbl>, COL2A1 <dbl>, COL3A1 <dbl>, COL4A1 <dbl>, COL4A2 <dbl>,
# COL4A3 <dbl>, COL4A3BP <dbl>, COL4A4 <dbl>, COL4A5 <dbl>, COL4A6 <dbl>,
# COL5A1 <dbl>, COL5A2 <dbl>, COL5A3 <dbl>, COL6A1 <dbl>, COL6A2 <dbl>,
# COL6A3 <dbl>, COL7A1 <dbl>, COL8A1 <dbl>, COL8A2 <dbl>, COL9A1 <dbl>,
# COL9A2 <dbl>, COL9A3 <dbl>These helpers make your code cleaner, more scalable, and easier to maintain. They can be used to select columns in tidyverse, also outside of across().
summarise() becomes more powerful!
So far, we’ve only applied a single function. But why stop at the mean? What if you want multiple statistics like mean, SD, min, and max - all in one go?
With across(), you can pass a list of functions:
df_comb %>%
summarise(across(.cols = starts_with("COL"),
.fns = list(mean, sd, min, max)))# A tibble: 1 × 136
COL10A1_1 COL10A1_2 COL10A1_3 COL10A1_4 COL11A1_1 COL11A1_2 COL11A1_3
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 5.20 1.78 2.79 10.3 6.38 2.62 2.76
# ℹ 129 more variables: COL11A1_4 <dbl>, COL11A2_1 <dbl>, COL11A2_2 <dbl>,
# COL11A2_3 <dbl>, COL11A2_4 <dbl>, COL13A1_1 <dbl>, COL13A1_2 <dbl>,
# COL13A1_3 <dbl>, COL13A1_4 <dbl>, COL14A1_1 <dbl>, COL14A1_2 <dbl>,
# COL14A1_3 <dbl>, COL14A1_4 <dbl>, COL15A1_1 <dbl>, COL15A1_2 <dbl>,
# COL15A1_3 <dbl>, COL15A1_4 <dbl>, COL16A1_1 <dbl>, COL16A1_2 <dbl>,
# COL16A1_3 <dbl>, COL16A1_4 <dbl>, COL17A1_1 <dbl>, COL17A1_2 <dbl>,
# COL17A1_3 <dbl>, COL17A1_4 <dbl>, COL18A1_1 <dbl>, COL18A1_2 <dbl>, …This gives you one wide row per column, with new columns like COL10A1_1, COL11A1_2, etc. A bit cryptic, right?
Let’s clean it up by naming the functions and columns:
gene_summary <- df_comb %>%
summarise(across(.cols = starts_with("COL"),
.fns = list(mean = mean,
sd = sd,
min = min,
max = max),
.names = "{.col}-{.fn}"))
gene_summary# A tibble: 1 × 136
`COL10A1-mean` `COL10A1-sd` `COL10A1-min` `COL10A1-max` `COL11A1-mean`
<dbl> <dbl> <dbl> <dbl> <dbl>
1 5.20 1.78 2.79 10.3 6.38
# ℹ 131 more variables: `COL11A1-sd` <dbl>, `COL11A1-min` <dbl>,
# `COL11A1-max` <dbl>, `COL11A2-mean` <dbl>, `COL11A2-sd` <dbl>,
# `COL11A2-min` <dbl>, `COL11A2-max` <dbl>, `COL13A1-mean` <dbl>,
# `COL13A1-sd` <dbl>, `COL13A1-min` <dbl>, `COL13A1-max` <dbl>,
# `COL14A1-mean` <dbl>, `COL14A1-sd` <dbl>, `COL14A1-min` <dbl>,
# `COL14A1-max` <dbl>, `COL15A1-mean` <dbl>, `COL15A1-sd` <dbl>,
# `COL15A1-min` <dbl>, `COL15A1-max` <dbl>, `COL16A1-mean` <dbl>, …Much better! Now the column names are readable and include both the variable and the statistic.
But still not your preferred format? You can probably pivot your way out of that!
gene_summary %>%
pivot_longer(cols = everything(),
names_to = c("gene", "statistic"),
names_sep = "-") %>%
pivot_wider(names_from = statistic,
values_from = value)# A tibble: 34 × 5
gene mean sd min max
<chr> <dbl> <dbl> <dbl> <dbl>
1 COL10A1 5.20 1.78 2.79 10.3
2 COL11A1 6.38 2.62 2.76 11.4
3 COL11A2 3.55 0.260 3.09 5.41
4 COL13A1 5.34 1.38 2.57 10.3
5 COL14A1 5.01 1.06 3.28 9.53
6 COL15A1 5.89 1.02 3.42 8.68
7 COL16A1 4.95 0.931 3.21 8.03
8 COL17A1 3.10 0.214 2.71 5.57
9 COL18A1 7.26 0.896 2.58 10.2
10 COL19A1 2.75 0.150 2.40 3.31
# ℹ 24 more rowsNow you get a long format table with one row per variable and all your stats in columns — clean, tidy, and ready for interpretation.
The anonymous function: ~ and .
We promised to get back to handling the NAs when doing summary stats. Let’s just add the na.rm=TRUE argument. To not have too many things going on at once we’ll only do mean() for now:
df_comb %>%
summarise(across(.cols = where(is.numeric),
.fns = list(mean = mean(na.rm = TRUE)),
.names = "{.col}-{.fn}"))Error in `summarise()`:
ℹ In argument: `across(...)`.
Caused by error in `mean.default()`:
! argument "x" is missing, with no defaultBrrrtt! We may not.
Why doesn’t this work?
When you pass functions directly into across() using the shorthand syntax (mean, sd, etc.), you’re only allowed to use the bare function with default arguments. You will also notice that we didn’t use brackets after their names, which is part of using the function shorthanded. Once you try to add something like na.rm = TRUE, the shorthandness breaks.
To pass arguments to a function that is called inside another function (i.e. calling mean inside summarise), we need to use what’s called an anonymous function. Don’t worry — it’s not as scary as it sounds.
It is written as a ~ and looks like this:
df_comb %>%
summarise(across(.cols = where(is.numeric),
.fns = list(mean = ~ mean(., na.rm = TRUE)),
.names = "{.col}-{.fn}"))# A tibble: 1 × 40
`age_at_initial_path_diagn-mean` days_to_tumor_recurren…¹ `days_to_death-mean`
<dbl> <dbl> <dbl>
1 59.7 624. 1010.
# ℹ abbreviated name: ¹`days_to_tumor_recurrence-mean`
# ℹ 37 more variables: `percent_normal_cells-mean` <dbl>,
# `percent_stromal_cells-mean` <dbl>, `percent_tumor_cells-mean` <dbl>,
# `COL10A1-mean` <dbl>, `COL11A1-mean` <dbl>, `COL11A2-mean` <dbl>,
# `COL13A1-mean` <dbl>, `COL14A1-mean` <dbl>, `COL15A1-mean` <dbl>,
# `COL16A1-mean` <dbl>, `COL17A1-mean` <dbl>, `COL18A1-mean` <dbl>,
# `COL19A1-mean` <dbl>, `COL1A1-mean` <dbl>, `COL1A2-mean` <dbl>, …Let’s break it down:
~to define the function.~ mean(., na.rm = TRUE)tells R:
“for each column, compute the mean, ignoring NAs”.is a placeholder for the current column being operated or ‘the data previously referred to’. We need to use the.becausemeanwhen called as a proper function needs to have an argument (a vector of numbers) to work on.
Multiple Summary Statistics with na.rm = TRUE
Now let’s compute several stats per column, all with na.rm = TRUE. Here we chose just the integer columns:
stats <- df_comb %>%
summarise(across(.cols = where(is.integer),
.fns = list(mean = ~ mean(., na.rm = TRUE),
sd = ~ sd(., na.rm = TRUE),
min = ~ min(., na.rm = TRUE),
max = ~ max(., na.rm = TRUE)),
.names = "{.col}-{.fn}")) %>%
# add reformating
pivot_longer(cols = everything(),
names_to = c("variable", "statistic"),
names_sep = "-") %>%
mutate(value = round(value, 1)) %>%
pivot_wider(names_from = statistic,
values_from = value)
print(stats)# A tibble: 6 × 5
variable mean sd min max
<chr> <dbl> <dbl> <dbl> <dbl>
1 age_at_initial_path_diagn 59.7 11.6 26 89
2 days_to_tumor_recurrence 624. 608. 8 5480
3 days_to_death 1010. 803. 8 5480
4 percent_normal_cells 2.4 6.7 0 55
5 percent_stromal_cells 12.8 11.9 0 70
6 percent_tumor_cells 80.6 15.4 0 100This produces a tidy table with one row per gene and columns for mean, sd, min, and max.
The helper functions can be used in other tidyverse operation such as mutate and select.
Handling Outliers
Now that we’ve explored summary statistics, it’s time to consider outliers — data points that differ markedly from the rest.
Let’s use our tidyverse skills to calculate some basic stats and visualize potential outliers.
stats <- stats %>%
mutate(thr_upper = mean + 2 * sd,
thr_lower = mean - 2 * sd) %>%
pivot_longer(cols = c(thr_lower, thr_upper), names_to = "thr_type", values_to = "threshold")
print(stats)# A tibble: 12 × 7
variable mean sd min max thr_type threshold
<chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 age_at_initial_path_diagn 59.7 11.6 26 89 thr_lower 36.5
2 age_at_initial_path_diagn 59.7 11.6 26 89 thr_upper 82.9
3 days_to_tumor_recurrence 624. 608. 8 5480 thr_lower -593.
4 days_to_tumor_recurrence 624. 608. 8 5480 thr_upper 1840.
5 days_to_death 1010. 803. 8 5480 thr_lower -596.
6 days_to_death 1010. 803. 8 5480 thr_upper 2615.
7 percent_normal_cells 2.4 6.7 0 55 thr_lower -11
8 percent_normal_cells 2.4 6.7 0 55 thr_upper 15.8
9 percent_stromal_cells 12.8 11.9 0 70 thr_lower -11
10 percent_stromal_cells 12.8 11.9 0 70 thr_upper 36.6
11 percent_tumor_cells 80.6 15.4 0 100 thr_lower 49.8
12 percent_tumor_cells 80.6 15.4 0 100 thr_upper 111. df_comb_longer <- df_comb %>%
pivot_longer(cols = where(is.integer),
names_to = "variable",
values_to = "value")
ggplot(df_comb_longer,
aes(x = value)) +
geom_histogram(bins = 30, fill="#482878FF") +
geom_vline(data = stats,
aes(xintercept = threshold),
color = "red", linetype = "dashed") +
facet_wrap(vars(variable), ncol = 3, scales = "free") +
theme_minimal()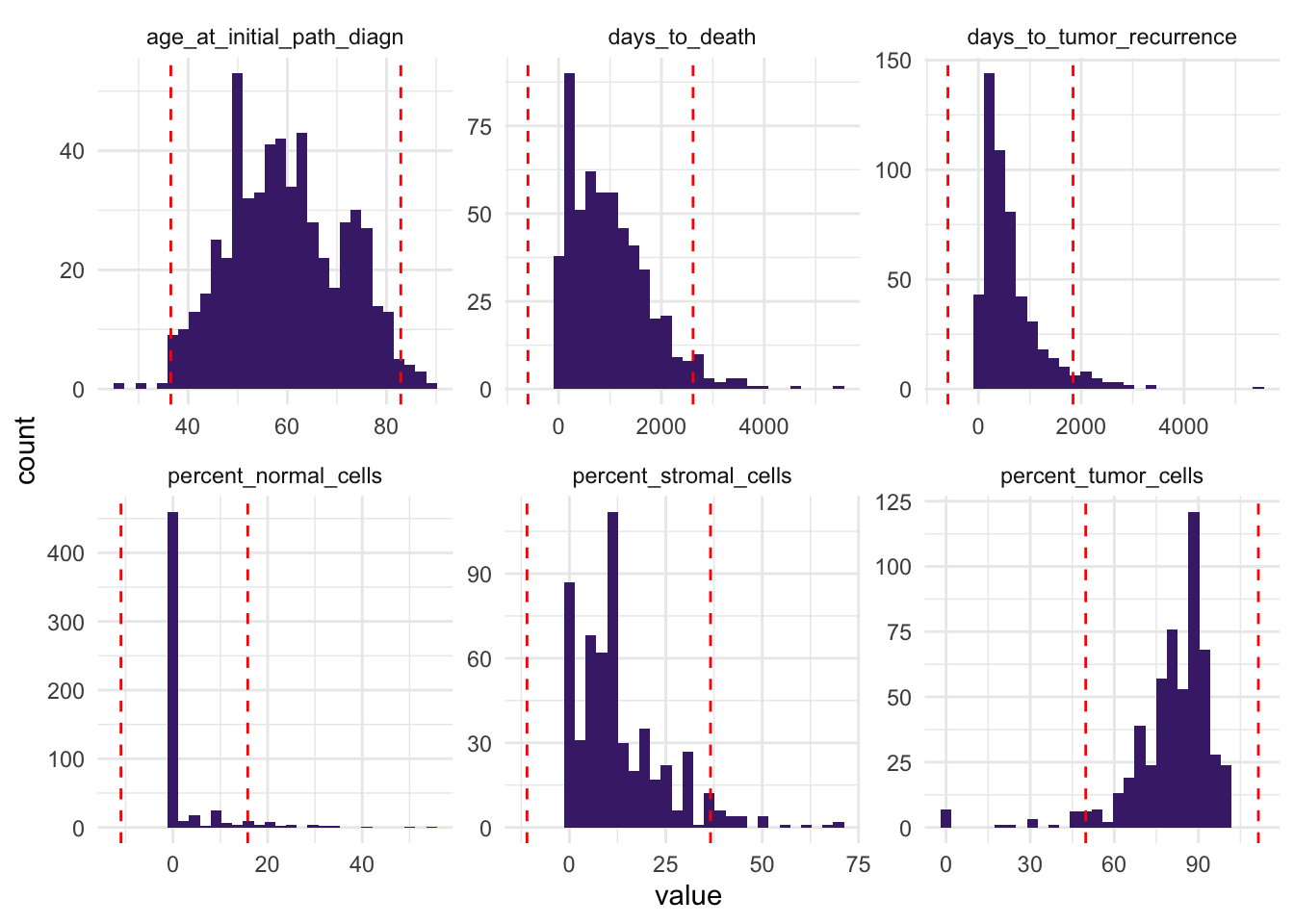
Sometimes, visualizing values in relation to other variables can make potential outliers easier to spot. Below, you’ll notice that a patient with 0% stromal cells and 0% tumor cells likely represents an outlier:
# Bivariate scatter plot colored by stromal cell percentage
ggplot(df_comb,
aes(x = percent_tumor_cells,
y = percent_stromal_cells,
color = percent_normal_cells)) +
geom_point() +
scale_color_viridis_c() +
theme_bw()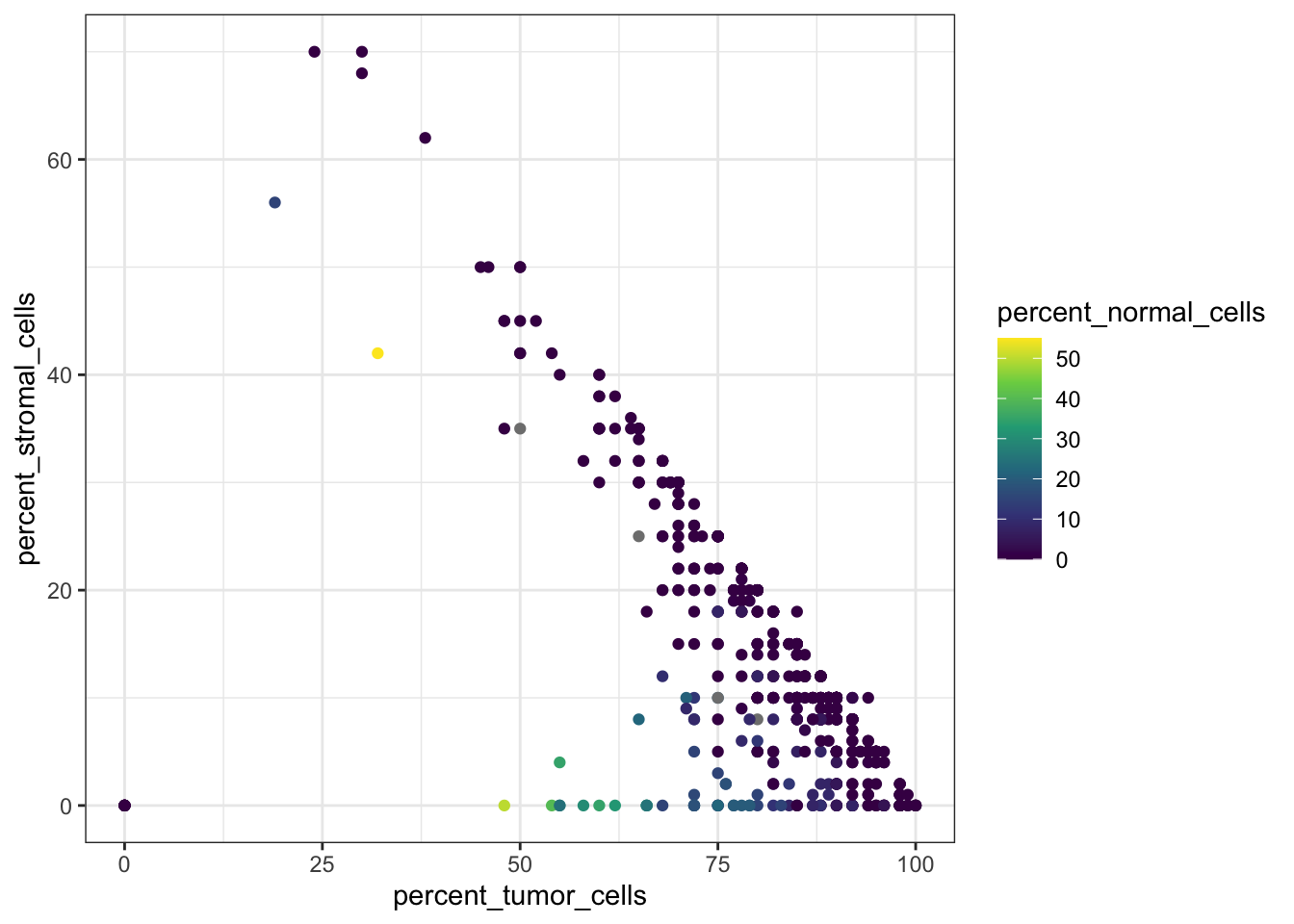
We probably want to remove patients where both percent_tumor_cells and percent_stromal_cells are 0%.
df_comb %>%
filter(percent_tumor_cells == 0 & percent_stromal_cells == 0)# A tibble: 7 × 53
unique_patient_ID sample_recode primarysite summarygrade tumorstage substage
<chr> <chr> <fct> <fct> <fct> <chr>
1 TCGA-24-1103 Tumor ov high 3 c
2 TCGA-23-1119 Tumor ov high 3 c
3 TCGA-23-1107 Tumor ov high 4 <NA>
4 TCGA-23-1120 Tumor ov high 3 c
5 TCGA-23-1121 Tumor ov high 3 c
6 TCGA-23-1123 Tumor ov high 3 c
7 TCGA-23-1124 Tumor ov high 3 c
# ℹ 47 more variables: grade <fct>, age_at_initial_path_diagn <int>,
# days_to_tumor_recurrence <int>, recurrence_status <fct>,
# days_to_death <int>, vital_status <fct>, percent_normal_cells <int>,
# percent_stromal_cells <int>, percent_tumor_cells <int>, batch <fct>,
# stage <fct>, dominant_cell_type <chr>, group <chr>, COL10A1 <dbl>,
# COL11A1 <dbl>, COL11A2 <dbl>, COL13A1 <dbl>, COL14A1 <dbl>, COL15A1 <dbl>,
# COL16A1 <dbl>, COL17A1 <dbl>, COL18A1 <dbl>, COL19A1 <dbl>, COL1A1 <dbl>, …df_comb <- df_comb %>%
filter(
(percent_tumor_cells != 0 | is.na(percent_tumor_cells) &
percent_stromal_cells != 0) | is.na(percent_stromal_cells))Sample vs variable outliers
There are many ways to detect and handle outliers. Instead of checking each variable separately, we can also look at all samples across multiple variables to spot unusual patterns.
One useful approach for datasets with many numeric or integer variables is to create a dendrogram using hierarchical clustering. This helps reveal samples that stand out from the rest.
Let´s try it, first, we select all integer columns and drop rows with missing values (they don’t help clustering).
df_int <- df_comb %>%
select(where(is.integer)) %>%
drop_na()
df_int %>% head()# A tibble: 6 × 6
age_at_initial_path_diagn days_to_tumor_recurrence days_to_death
<int> <int> <int>
1 61 442 701
2 60 574 574
3 53 428 1264
4 78 61 61
5 74 870 789
6 73 68 84
# ℹ 3 more variables: percent_normal_cells <int>, percent_stromal_cells <int>,
# percent_tumor_cells <int>Next, we scale the data (details in Presentation 3), calculate pairwise distances between all pairs of scaled variables, and run hierarchical clustering to build a dendrogram.
This tree-like plot helps reveal samples that cluster far from the rest — potential outliers.
# Euclidean pairwise distances
df_int_dist <- df_int %>%
mutate(across(everything(), scale)) %>%
dist(., method = 'euclidean')
# Hierarchical clustering with Ward's distance metric
hclust_int <- hclust(df_int_dist, method = 'average')If the plot is hard to see:
Copy-paste the code into the console to open it in the Plots pane.
Click Export → Copy to Clipboard… and enlarge it as needed.
Or save it as a high-resolution file and inspect it outside RStudio.
# Plot the result of clustering
# png("dendrogram_plot.png", width = 2000, height = 500)
# plot(hclust_int, main = "Clustering based on scaled integer values", cex = 0.7)
# dev.off()
fviz_dend(hclust_int, h = 10)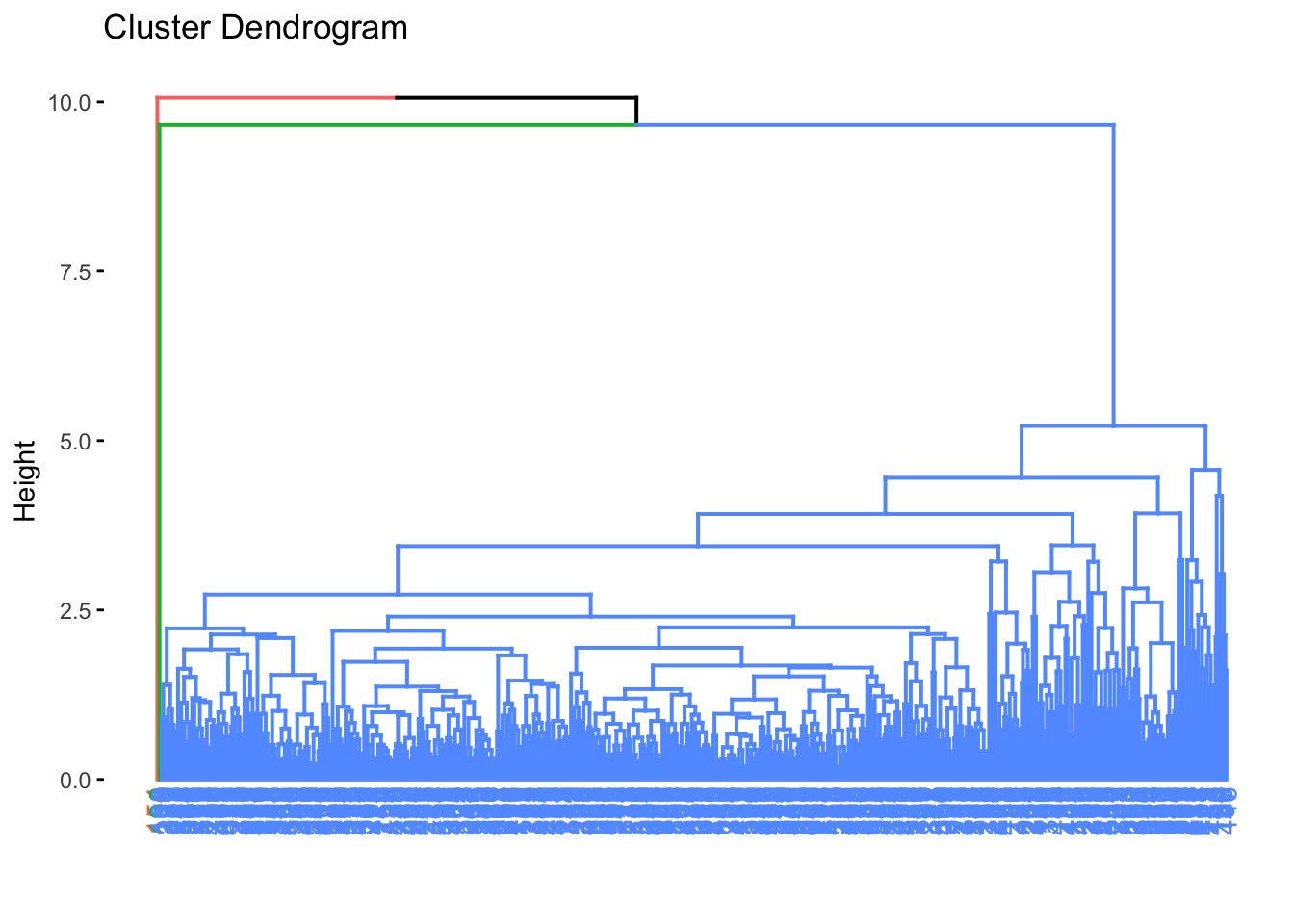
From the dendrogram, we can see a few samples that branch off far from the main cluster — these long branch lengths (Height) may indicate outliers.
Let’s identify samples with unusually large heights and inspect them:
# Set a high threshold (top 0.1%)
#threshold <- quantile(hclust_int$height, 0.99)
threshold <- 6
# Find their order in the clustering
ord_inx <- hclust_int$order[hclust_int$height > threshold]
ord_inx[1] 180 97# See which samples they are
df_int %>% slice(ord_inx)# A tibble: 2 × 6
age_at_initial_path_diagn days_to_tumor_recurrence days_to_death
<int> <int> <int>
1 56 213 1354
2 51 192 192
# ℹ 3 more variables: percent_normal_cells <int>, percent_stromal_cells <int>,
# percent_tumor_cells <int>Next, we’ll bring back our summary statistics table to see whether these measurements fall outside what’s typical for the dataset.
stats# A tibble: 12 × 7
variable mean sd min max thr_type threshold
<chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 age_at_initial_path_diagn 59.7 11.6 26 89 thr_lower 36.5
2 age_at_initial_path_diagn 59.7 11.6 26 89 thr_upper 82.9
3 days_to_tumor_recurrence 624. 608. 8 5480 thr_lower -593.
4 days_to_tumor_recurrence 624. 608. 8 5480 thr_upper 1840.
5 days_to_death 1010. 803. 8 5480 thr_lower -596.
6 days_to_death 1010. 803. 8 5480 thr_upper 2615.
7 percent_normal_cells 2.4 6.7 0 55 thr_lower -11
8 percent_normal_cells 2.4 6.7 0 55 thr_upper 15.8
9 percent_stromal_cells 12.8 11.9 0 70 thr_lower -11
10 percent_stromal_cells 12.8 11.9 0 70 thr_upper 36.6
11 percent_tumor_cells 80.6 15.4 0 100 thr_lower 49.8
12 percent_tumor_cells 80.6 15.4 0 100 thr_upper 111. Now we can compare these flagged samples to our summary statistics to understand why they stand out - and decide if they should be removed.
For these samples:
Age at diagnosis and days to death are within a normal range.
However, percent stromal cells and percent tumor cells are unusually low.
These samples have uncharacteristically high normal cell proportions, which is extreme for this dataset.
Given that they show extreme values across several key variables, it’s reasonable to treat them as outliers and remove them to avoid bias in our analysis.
df_comb <- df_comb %>%
filter(!(
(percent_normal_cells == 35 & days_to_death == 1354) |
(percent_normal_cells == 30 & days_to_death == 192)
) |
is.na(percent_normal_cells) | is.na(days_to_death))Be cautious when removing data: Outlier removal should always be guided by domain knowledge and clear justification. Removing too much — or for the wrong reasons — can distort your analysis.
Handling Missing Data
Missing data is common in most real-world datasets and can significantly affect the quality of our analysis. During EDA, it’s essential to identify, understand, and properly handle missing values to avoid biased or misleading conclusions.
Let’s inspect rows with the most missing values:
df_comb %>%
mutate(na_count = rowSums(is.na(.))) %>%
arrange(desc(na_count)) %>%
slice_head(n = 10)# A tibble: 10 × 54
unique_patient_ID sample_recode primarysite summarygrade tumorstage substage
<chr> <chr> <fct> <fct> <fct> <chr>
1 TCGA-01-0630 Healthy <NA> <NA> <NA> <NA>
2 TCGA-01-0631 Healthy <NA> <NA> <NA> <NA>
3 TCGA-01-0633 Healthy <NA> <NA> <NA> <NA>
4 TCGA-01-0636 Healthy <NA> <NA> <NA> <NA>
5 TCGA-01-0637 Healthy <NA> <NA> <NA> <NA>
6 TCGA-01-0628 Healthy <NA> <NA> <NA> <NA>
7 TCGA-01-0639 Healthy <NA> <NA> <NA> <NA>
8 TCGA-01-0642 Healthy <NA> <NA> <NA> <NA>
9 TCGA-04-1353 Tumor <NA> <NA> <NA> <NA>
10 TCGA-36-2539 Tumor <NA> <NA> <NA> <NA>
# ℹ 48 more variables: grade <fct>, age_at_initial_path_diagn <int>,
# days_to_tumor_recurrence <int>, recurrence_status <fct>,
# days_to_death <int>, vital_status <fct>, percent_normal_cells <int>,
# percent_stromal_cells <int>, percent_tumor_cells <int>, batch <fct>,
# stage <fct>, dominant_cell_type <chr>, group <chr>, COL10A1 <dbl>,
# COL11A1 <dbl>, COL11A2 <dbl>, COL13A1 <dbl>, COL14A1 <dbl>, COL15A1 <dbl>,
# COL16A1 <dbl>, COL17A1 <dbl>, COL18A1 <dbl>, COL19A1 <dbl>, COL1A1 <dbl>, …Looking at our missing values, we see that most missingness comes from samples labeled as Healthy(sample_recode).
This makes sense: Healthy tissue lacks tumor-specific details like tumor stage or percent tumor cells. This is an example of MAR (Missing At Random) — the missingness is related to an observed category (healthy vs. tumor tissue).
Since our focus is on tumor samples, not healthy tissue (few samples), we can remove these samples to clean up our data and avoid noise.
df_comb <- df_comb %>% filter(sample_recode == "Tumor")
# Let’s inspect rows with the most missing values again.
df_comb %>%
mutate(na_count = rowSums(is.na(.))) %>%
arrange(desc(na_count)) %>%
slice_head(n = 10)# A tibble: 10 × 54
unique_patient_ID sample_recode primarysite summarygrade tumorstage substage
<chr> <chr> <fct> <fct> <fct> <chr>
1 TCGA-04-1353 Tumor <NA> <NA> <NA> <NA>
2 TCGA-36-2539 Tumor <NA> <NA> <NA> <NA>
3 TCGA-42-2593 Tumor ov <NA> <NA> <NA>
4 TCGA-04-1341 Tumor ov high <NA> <NA>
5 TCGA-57-1994 Tumor ov <NA> <NA> <NA>
6 TCGA-24-1104 Tumor ov high 4 <NA>
7 TCGA-23-1118 Tumor ov high 3 c
8 TCGA-24-1105 Tumor ov high 3 c
9 TCGA-23-1113 Tumor ov high 4 <NA>
10 TCGA-13-0755 Tumor ov high 4 <NA>
# ℹ 48 more variables: grade <fct>, age_at_initial_path_diagn <int>,
# days_to_tumor_recurrence <int>, recurrence_status <fct>,
# days_to_death <int>, vital_status <fct>, percent_normal_cells <int>,
# percent_stromal_cells <int>, percent_tumor_cells <int>, batch <fct>,
# stage <fct>, dominant_cell_type <chr>, group <chr>, COL10A1 <dbl>,
# COL11A1 <dbl>, COL11A2 <dbl>, COL13A1 <dbl>, COL14A1 <dbl>, COL15A1 <dbl>,
# COL16A1 <dbl>, COL17A1 <dbl>, COL18A1 <dbl>, COL19A1 <dbl>, COL1A1 <dbl>, …There are two main approaches when deciding what to do:
Remove rows with missing values
Impute values
In our case:
We’ll replace numeric missing values with the median of each variable. We’ll replace missing factor levels with the most common category.
df_comb <- df_comb %>%
mutate(across(
c(
percent_normal_cells,
percent_stromal_cells,
percent_tumor_cells,
age_at_initial_path_diagn,
days_to_death,
days_to_tumor_recurrence
),
~ if_else(is.na(.x), as.integer(median(.x, na.rm = TRUE)), as.integer(.x))
))
df_comb <- df_comb %>%
mutate(across(
c(stage, grade),
~ fct_explicit_na(.x, na_level = names(sort(table(.x), decreasing = TRUE))[1])
))Warning: There was 1 warning in `mutate()`.
ℹ In argument: `across(...)`.
Caused by warning:
! `fct_explicit_na()` was deprecated in forcats 1.0.0.
ℹ Please use `fct_na_value_to_level()` instead.We’ll keep the remaining NAs as-is for now.
However always consider the impact of missing data. Even after imputing, missing data can cause uncertainty and bias so understands the result with caution.
In Summary
We conducted an initial exploratory data analysis (EDA) and, overall, the dataset looks clean and well-structured.
Reshaped data — switched between wide and long formats to easily plot multiple variables together.
Checked factors — reviewed category balance, merged sparse groups, and visualized how factors relate.
Generated and inspected summary statistics — used tidyverse helpers to calculate means, standard deviations, and more for many columns at once, confirming that values align with biological expectations.
Detected and removed outliers — used clustering to identify and exclude extreme or inconsistent samples with unusually high normal cell content.
Handled missing data thoughtfully — removed irrelevant samples and imputed remaining gaps where appropriate.
Key Takeaways
Long format is your best friend when plotting or modeling.
Use
across()and helpers to write clean, scalable summaries.Visual checks are as important as statistical checks.
EDA is not optional — it’s the foundation of trustworthy analysis.
Keep your workflow reproducible and tidy for future modeling stages.
save(df_comb, file="../data/Ovarian_comb_prep_Col.RData")