Pandas¶
Introduction¶
Popular package for data science: offers powerful data structures that make data manipulation and analysis easy.
The DataFrame is one of them.
Pandas is built on top of numpy.
Pandas is well suited for tabular data with heterogeneously-typed columns, as in an Excel spreadsheet
Has an interface to directly plot using maptlotlib, seaborn, plotly, for example.
Two main classes (types/ objects)¶
pandas.Seriespandas.DataFrame
- a
Seriesis anumpy.arraywith anIndexseries. - a column in a
DataFrameis aSeries. - columns in a
DataFrameshare anIndex
Import relevant packages¶
import pandas as pd
Creating an instance of a pandas.Series¶
There are many ways. E.g from a list, a built-in range and numpy arrays.
But a Series is an object holding some data.
Let's create a Series from a list and a built-in range.
series_list = pd.Series([3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
series_list
series_range = pd.Series(range(3, 13))
series_range
print("Series from list, underlying data:\t", series_list.values)
print("Series from range, underlying data:\t", series_range.values)
series_list.values
series_range.values
Difference: indexing of data¶
- pandas names "data" by an row indices (and column indices for 2D structures)
- see also recent talk on PyData2021 by James Powell (it's relatively fast)
series1 = pd.Series([1,2,3], index=['row1', 'row2', 'row3'])
series1
series2 = pd.Series([1,2,4])
series2.index = ['row1', 'row2', 'row4']
series2
series1 + series2
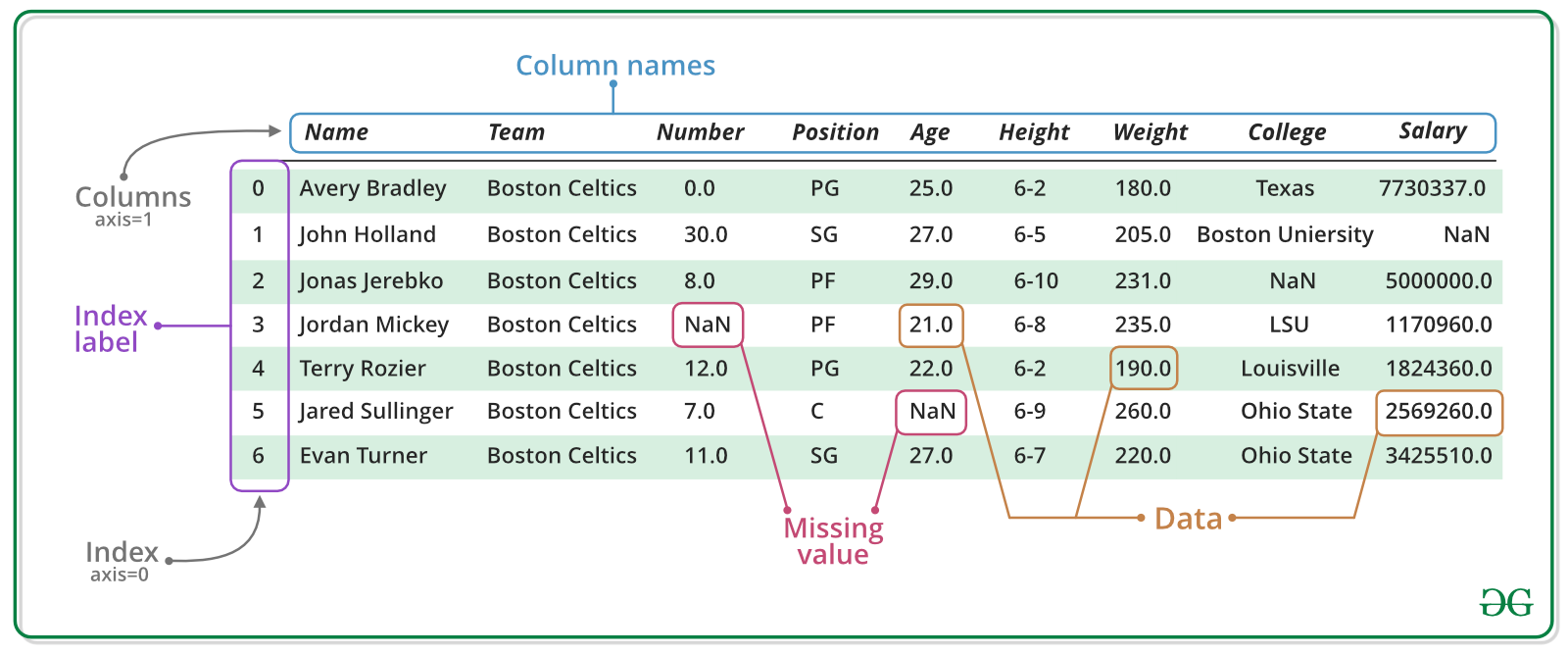
What is a DataFrame?¶
A DataFrame is basically, a Table of data (or a tabular data structure) with labeled rows and columns. The rows are labeled by a special data structure called an Index, that permits fast look-up and powerful relational operations.
Create a DataFrame directly¶
From a list of lists¶
data = [
[2.23, 1, "test"],
[3.45, 2, "train"],
[4.5, 3, "test"],
[6.0, 4, "train"]
]
df = pd.DataFrame(data, columns=['A', 'B', 'C'])
df
From a list of dicts¶
data = [
{'A': 2.23, 'B': 1, 'C': "test"},
{'A': 3.45, 'B': 2, 'C': "train"},
{'A': 4.5, 'B': 3, 'C': "test"},
{'A': 6.0, 'B': 4, 'C': "train"}
]
df = pd.DataFrame(data)
df
From a Dict of Lists¶
data = {
"A": [2.23, 3.45, 4.5, 6.0],
"B": [1, 2, 3, 4],
"C": ["test", "train", "test", "train"],
}
df = pd.DataFrame(data)
df
From a dict of dicts¶
data = {
"row1": {"A": 2.23, "B": 1, "C": "test"},
"row3": {"A": 3.45, "B": 2, "C": "train"},
"row2": {"A": 4.5, "B": 3, "C": "test"},
"row4": {"A": 6.0, "B": 4, "C": "train"},
}
df = pd.DataFrame.from_dict(
data,
orient="index", # default is columns. pd.DataFrame also works, but you have to transpose the data
)
df
From an empty DataFrame¶
df = pd.DataFrame()
df['A'] = [2.23, 3.45, 4.5, 6.0]
df['B'] = [1, 2, 3, 4]
df['C'] = ["test", "train", "test", "train"]
df
Indexing of data¶
data = [
[2.23, 1, "test"],
[3.45, 2, "train"],
[4.5, 3, "test"],
[6.0, 4, "train"]
]
df = pd.DataFrame(data, index=['row1', 'row2', 'row3', 'row4'], columns=['A', 'B', 'C'])
df
df.index = ['a', 'b', 'c', 'd']
df
Exercise 1¶
Please recreate the table below as a Dataframe using one of the approaches detailed above:
| Year | Product | Cost | |
|---|---|---|---|
| 0 | 2015 | Apples | 0.35 |
| 1 | 2016 | Apples | 0.45 |
| 2 | 2015 | Bananas | 0.75 |
| 3 | 2016 | Bananas | 1.10 |
Which approach did you prefer? Why?
Loading data into DataFrames from a file¶
Pandas has functions that can make DataFrames from a wide variety of file types. To do this, use one of the functions in Pandas that start with read_. Here is a non-exclusive list of examples:
| File Type | Function Name |
|---|---|
| Excel | pd.read_excel |
| CSV, TSV | pd.read_csv |
| H5, HDF, HDF5 | pd.read_hdf |
| JSON | pd.read_json |
| SQL | pd.read_sql_table |
These are all functions, which can be called, i.e.
pd.read_csv()
Loading the Data¶
The file can be local or hosted: The read_*-function have many options and are very high general (in the sense of broad or comprehensive) functions.
url_ecdc_daily_cases = "https://opendata.ecdc.europa.eu/covid19/nationalcasedeath_eueea_daily_ei/csv/data.csv"
df = pd.read_csv(url_ecdc_daily_cases)
df
Examining the Dataset¶
Sometimes, we might just want to quickly inspect the DataFrame:
Attributes¶
df.shape # Shape of the object (2D)
df.dtypes # Data types in each column
df.index # Index range
df.columns # Column names
Functions¶
df.head() # Displays first 5 rows
df.tail() # Displays last 5 rows
df.sample() # Displays randow 5 rows
df.info() # DataFrame information
Shape¶
The first dimension are the number of rows (the length of the DataFrame), the second dimension the number of features or columns. The direction going down the rows is axis=0 or axis='index', and going over the columns is axis=1 or axis='columns'.
| axis | descriptions |
|---|---|
| 0 | index |
| 1 | columns |
df.shape
Data types¶
df.dtypes
Index and Columns¶
df.index
df.columns
Head, tail and sample¶
df.head()
df.tail()
df.sample(15)
Info¶
df.info(memory_usage='deep')
_ = df.info()
print(_)
Renaming index/column names¶
Data might come to us with index names, column names or othe naming conventions that do not fit our requirements.
You can change those names to something more fitting, using the rename() function.
If you want to change index names and/or column names, use the keyword index or columns, respectively, and pass a dictionary with the original index/column name as key, and the new name as value.
df.rename(columns={'A':'Column A', 'D':'Column D'})
df.rename(index={0:'row1', 1:'row2', 100:'end row'})
You can also pass functions to rename():
df.rename(index=str) # Change the index data type to string
df.rename(columns=str.lower) # Make all column names lowercase
To keep the changes, set the inplace argument to True, or store the changed DataFrame in a variable:
df.rename(index=str, inplace=True)
new_df = df.rename(index=str)
rename_df = df.rename(columns={'day':'Column day', 'deaths':'Column deaths'}, index={0:'row1', 1:'row2', 100:'end row'})
rename_df.head()
Indexing and Selecting Data¶
Native accessors¶
Pandas has a lot of flexibility in the number of syntaxes it supports. For example, to select columns in a DataFrame:
df['Column1']
df.Column1 # no whitespaces possible!
Multiple Columns can also be selected by providing a list:
df[['Column1', 'Column2']]
df['day']
df[['year', 'month', 'day']]
df['day'][0]
Pandas accessors¶
Accessor operators iloc (index-based) and loc (label-based):
df.iloc[5] # df.iloc[row_index, column_index]
df.loc['Row6'] # df.loc[row_label, column_label]
On its own, the ":" operator means "everything".
When combined with other selectors, can be used to indicate a range of values.
In loc, both start bound and the stop bound are inclusive, while in iloc, the stop bound is exclusive.
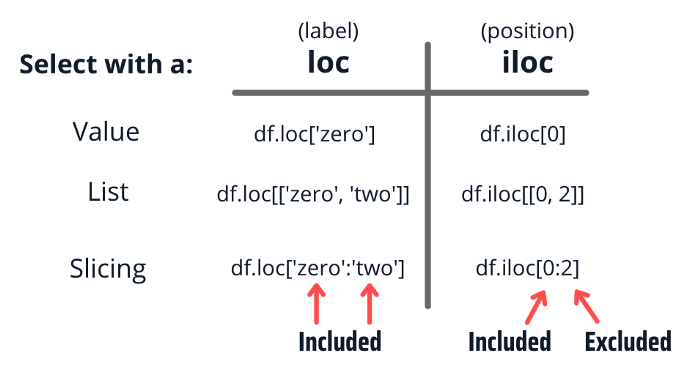
Index-based selection¶
Select an entire column:
df.iloc[:, 5] # all rows, and 6th column
Select the first 3 rows of the 6th column:
df.iloc[:3, 5]
Select only the second and third rows:
df.iloc[1:3, 5]
Select with a list:
df.iloc[[2, 3, 4, 5], 5]
Use negative numbers:
df.iloc[-5:]
Label-based selection¶
df.loc[:, 'deaths']
df.loc[:2, 'deaths']
df.loc[1:2, 'deaths']
df.loc[[2, 3, 4, 5], 'deaths']
df.loc[22828:,['dateRep', 'cases', 'deaths', 'countriesAndTerritories']]
Modifying the index¶
In small DataFrames:
df = pd.DataFrame(data, index=['row1', 'row2', 'row3', 'row4'], columns=['A', 'B', 'C'])
or
df.index = ['row1', 'row2', 'row3', 'row4']
But in big DataFrames there are too many rows to make the above efficient. So we use set_index():
df.set_index('Column1')
To keep the index change, store it as a variable:
df = df.set_index('Column1')
or use the argument inplace=True:
df.set_index('Column1', inplace=True)
new_df = df.set_index('countriesAndTerritories')
new_df.head()
new_df.loc['Denmark']
Exercise 2¶
Display the first 5 lines of the dataset.
Show the last 15 lines
Check 10 random lines of the dataset
Make a new dataframe containing just the date, population data and number of cases and deaths
Make a new dataframe containing just the 10th, 15th and 16th lines of the dataset. Which method did you use? Why?
Conditional selection¶
Row selection¶
Defining a condition with a Pandas DataFrame follows the same syntax:
df['Column1'] > 0 # Return a Series of True and False
To filter the DataFrame for the rows where the value is True:
select_rows = df[df['Column1'] > 0]
or
select_rows = df.loc[df['Column1'] > 0]
You can also filter based on multiple conditions, using the element-wise ("bit-wise") logical operators & data intersection, or | for the data union.
select_rows = df[(df['Column1'] > 0) & (df['Column2'] > 2)]
or
select_rows = df.loc[(df['Column1'] > 0) & (df['Column2'] > 2)]
select_rows = df[(df['Column1'] > 0) | (df['Column2'] > 2)]
or
select_rows = df.loc[(df['Column1'] > 0) | (df['Column2'] > 2)]
Consider first creating the mask (Series of True and False values indicating if a row is selected)
mask = (df['Column1'] > 0) | (df['Column2'] > 2)
select_rows = df[mask]
Row and Column selection¶
select_df = df.loc[df['Column1'] > 0, ['Column1', 'Column2', 'Column3']]
or
select_df = df[df['Column1'] > 0][['Column1', 'Column2', 'Column3']]
or
select_df = df[['Column1', 'Column2', 'Column3']][df['Column1'] > 0]
Using masks:
row_mask = (df['Column1'] > 0) | (df['Column2'] > 2)
column_mask = ['Column4', 'Column5']
select_df = df.loc[row_mask, column_mask]
Checkout more interesting methods:
df.loc[df['countriesAndTerritories'] == 'Denmark'] # This is not saved
filtered_df = df.loc[df['countriesAndTerritories']=='Denmark', ['dateRep', 'cases']]
filtered_df.head()
Exercise 3¶
Make a new dataframe with the year, month, day, number of cases and deaths, for countries in Europe.
With what you learned so far, how would you calculate the daily deaths, on average, for Austria?
We will solve together
Summary functions¶
Pandas' Series and DataFrames are iterables, and can be given to any function that expects a list or Numpy Array, which allows them to be useful to many different libraries' functions. For example, to compute basic statistics for a colum (Series):
df.describe() # describe numeric columns
df['Column1'].describe() # describe a particular column/series
df['Column1'].count() # counts non-NA cells
df['Column1'].nunique() # counts number of distinct elements in specified axis
df['Column1'].unique() # returns array of unique values of Series, in order of appearance
df['Column1'].value_counts() # returns list of unique values and how often they occur in the dataset
df['Column1'].max()
df['Column1'].mean()
df['Column1'].idxmax()
or for row:
df.loc['row_index_label'].sum() # count, std, mean, etc
or for all columns
df.mean() # default by column (= over all index)
or for all rows
df.mean(axis=1)
df.mean(axis='columns') # columns axis is axis 1
What the default axis for a method (or operation) will vary.
Example documentation: var
df.mean() # uses numeric column only
Exercise 4¶
Which country has the maximum number of deaths reported on one day?
Hint: check Pandas DataFrame API reference for more useful functions
How many countries does europe have?
How many unique dates are in this data set?
What is the average daily deaths for Norway?
Modifying Data¶
Any transformation function can be performed on each element of a column or on the entire DataFrame.
You can assign a constant value:
df['Column20'] = 10
An iterable of values:
df['Column21'] = range(len(df), 0, -1) # Replace the entire column with other values (length must match)
You can create a columns based on the transformation of another columns' numerical values:
df['Column22'] = df['Column1'] * 5
Or modify strings:
df['Column10'] = df['Column10'].str.upper()
Or even delete an entire column:
del df['B']
Column: a
pandas.Serieswith the index of the rows
# Create new column based on the number of cases per 100.000 population
df['cases_per_100k'] = df['cases'] / df['popData2020'] * 100000
# Filter dataset to include only Denmark
mask_denmark = df['countriesAndTerritories'] == 'Denmark'
df[mask_denmark]
For more complicated operations, where you want to combine DataFrames with Series, you can have a look how broadcasting works in pandas.
Exercise 5¶
Which country has most daily deaths per 100.000?
Which country, and on what date, had the lowest number of infected in Europe?
Make the country code variable column lower-cased
Make a column called "alive", to be the opposite of the deaths column
GroupBy Operations and Sorting¶
In most of our tasks, getting single metrics from a dataset is not enough, and we often actually want to compare metrics between groups or conditions.
The groupby method essentially splits the data into different groups depending on a variable of your choice, and allows you to apply summary functions on each group. For example, if you wanted to calculate the mean number of cases by month from a given our DataFrame:
df.groupby('month').cases.mean()
where "month" is a column name from the DataFrame.
You can also group by multiple columns, by providing a list of column names:
df.groupby(['year', 'month']).cases.mean()
Aggregating by multiple columns will create an
MultiIndexrow-Index.
You can access indices with more than one entry using tuples:df.loc[(first_index, second_index)]
The groupby method returns a GroupBy object, where the .groups attribute is a dictionary whose keys are the computed unique groups.
GroupBy objects are lazy, meaning they don't start calculating anything until they know the full pipeline. This approach is called the "Split-Apply-Combine" workflow. You can get more info on it in the UserGuide.
df.groupby(by='countriesAndTerritories').cases.sum().loc['Denmark']
df.loc[mask_denmark, 'cases'].sum()
Exercise 6¶
What was the median number of daily cases per country?
How many days where there without deaths in each country?
- select rows with zero deaths, group these and count the days
How many infected daily on average for each country in each month?
What was the infection rate for each country for the whole period?
For the purposes of this exercise, consider infection rate is the total number of infected divided by the population size.
Multiple Statistics per Group¶
Another piece of syntax we are going to look at, is the aggregate method for a GroupBy pandas objects, also available as agg.
The aggregation functionality provided by this function allows multiple statistics to be calculated per group in one calculation.
The instructions to the method agg are provided in the form of a dictionary, where the keys specify the columns upon which to apply the operation(s), and the value specify the function to run:
df.groupby(['year', 'month']).agg({'duration':'sum',
'network_type':'count',
'date':'first'})
Not a working example for our
DataFrame
You can also apply multiple functions to one column in groups:
df.groupby(['year', 'month']).agg({'duration':[min, max, sum],
'network_type':'count',
'date':[min, 'first', 'nunique']})
Aggregating by multiple columns will create an
MultiIndexrow-Index.
You can access indices with more than one entry using tuples:df.loc[(first_index, second_index)]
new_df = df.groupby(by='countriesAndTerritories').agg({'cases': 'sum', 'popData2020': 'mean'})
new_df['cases']/new_df['popData2020']
Exercise 7¶
Calculate how many were infected and how many died daily, on average, in each country in Europe?
What was the highest and the lowest number of infected per month, per country?
Sorting¶
Sort a dataset based on column values, with sort_values():
df.sort_values(by='cases')
By default, values are sorted in ascending order (alhpabetically). To change to descending order, change the argument ascending to False:
df.sort_values(by='cases', ascending=False)
To sort the dataset based on index values, use the companion function sort_index(). This function has the same arguments and default order:
df.groupby(by='countriesAndTerritories').agg({'cases': 'sum', 'deaths': sum}).sort_index()
You can also sort by more than one column at a time:
df.sort_values(by=['countriesAndTerritories', 'cases'], ascending=False)
df.sort_values('cases_per_100k', ascending=False)
Extra: Handling Missing Values¶
Missing values are often a concern in data science, for example in proteomics, and can be indicated with a None or NaN (np.nan in Numpy). Pandas DataFrames have several methods for detecting, removing and replacing these values:
| method | description |
|---|---|
isna() | Returns True for each NaN | notna() | Returns False for each NaN | dropna() | Returns just the rows without any NaNs |
Detect missing values and retrieve rows where they are present in column "Column1":
missing_data_rows = df[df['Column1'].isna()]
Imputation¶
Imputation means replacing the missing values with real values.
| method | description |
|---|---|
fillna() | Replaces the NaNs with values (provides lots of options) |
ffill() | Replaces the Nans with the previous non-NaN value (equivalent to df.fillna(method='ffill') |
bfill() | Replaces the Nans with the following non-NaN value (equivalent to df.fillna(method='bfill') |
interpolate() | interpolates nans with previous and following values |
Replace missing values with constant value across dataset:
df = df.fillna(0)
Replace missing values in a specific column:
df['Column1'] = df['Column1'].fillna(0)
Replace missing values with specific values per column, using a dictionary:
new_values = {'Column1': 0, 'Column2': 5, 'Column3': 'Unknown'}
df = df.fillna(values=new_values)
Exercise 8¶
Here we will use the titanic data which contains some missing values
url = 'https://raw.githubusercontent.com/mwaskom/seaborn-data/master/titanic.csv'
titanic = pd.read_csv(url)
titanic
What proportion of the "deck" column is missing data?
How many rows don't contain any missing data at all?
Make a dataframe with only the rows containing no missing data.
Exercise 9¶
Using the following DataFrame, solve the exercises below.
recreate it in every exercise or copy it to avoid confusion:
data_type_filled = data.copy()
Can you explain the problem?
data = pd.DataFrame({'time': [0.5, 1., 1.5, None, 2.5, 3., 3.5, None], 'value': [
6, 4, 5, 8, None, 10, 11, None]})
data
Replace all the missing "value" rows with zeros.
Replace the missing "time" rows with the previous value.
Replace all of the missing values with the data from the next row. What do you notice when you do this with this dataset?
Linearly interpolate the missing data. What is the result for this dataset?
Advanced material¶
Wide vs Long formats¶
Two formats for Pandas DataFrames: wide and long.
In the wide format, each feature (attribute) is a separate columns, while each row represents many features of the same individual entry. In the wide format, there are no repeated records, but there might be missing values. This format is preferable to perform statistics (e.g. mean).
In the long format, each row only shows one feature for each individual entry, and there are multiple rows for each entry (one for each feature). We often use this format for graphic plotting.
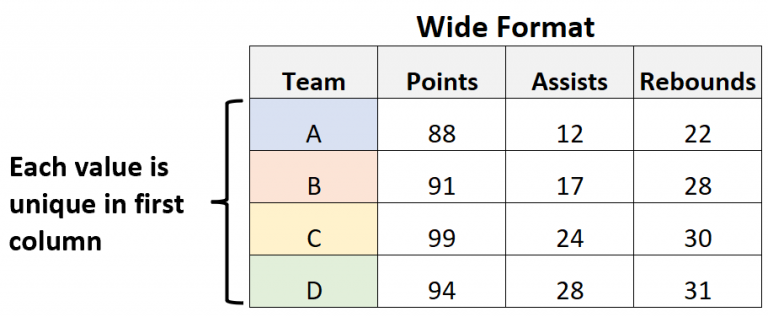 | 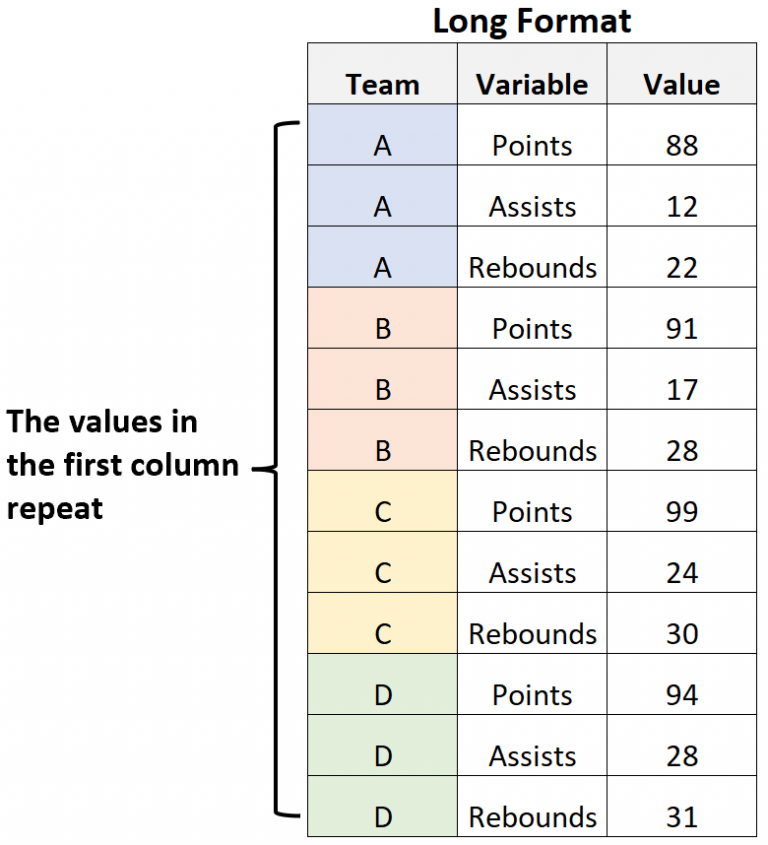 |
Lambda and Mapping functions¶
You can, of course, write your own functions in Python, but lambda functions are often a faster and easier way to write simple functions on the fly.
To do so, you always start with the lambda keyword, followed by the names of the arguments, a colon and then the expression than specifies what we want the function to return. For example, if we want multiply two numbers:
max_value = lambda x, y: x * y
a = 55
b = 34
max_value(a, b)
lambda functions are especially useful when combined with mapping methods like map() and apply().
The map term indicates a function that takes one set of values and "maps" them to another set of values. In data science, we often need to create new representations or transform existing data. mapping functions help us do this work.
map() is slightly simpler and takes in a function as argument, for example, a lambda function. Whatever function you pass though, make sure the expexted input is always a single value. The return of map() will be the original input value, transformed by the lambda function.
average_cases = df.cases.mean()
df.cases.map(lambda x: x - average_cases) # Centers the data distribution of number of cases around 0.
Besides functions, map() can also accept dictionaries (key corresponds to input value, and dictionary value is the new value to replace the input one), and Series.
apply() is the equivalent method but when we want to transform an entire DataFrame. It can be applied row-wise or column-wise, depending whether the argument axis is set to rows or columns, respectively.
average_cases = df.cases.mean()
def center_mean(data):
data.cases = data.cases - average_cases
return data
df.apply(center_mean, axis='columns')
Differently from map(), apply() also allows passing of positional or keyword arguments to the function.
def get_deaths_class(value, lower_threshold, upper_threshold):
if value >= int(upper_threshold):
class_name = 'High'
elif value <= int(lower_threshold):
class_name = 'Low'
else:
class_name = 'Moderate'
return class_name
df['deaths_class'] = df['deaths'].apply(get_deaths_class, lower_threshold = 20, upper_threshold = 100)
You can also use the apply() method in a group-wise analysis:
df.groupby(['countriesAndTerritories', 'year', 'month']).apply(lambda df: df.loc[df.cases.idxmax()])
For more information of these methods and more, go to Pandas API reference
Combining DataFrames¶
When performing operations on a dataset, we might sometimes need to combine different DataFrames and/or other Series. Pandas has a few functions that allows to do that: concat(), join(), and merge().
concat() is the simplest combining methods. It will smush together all the elements in a list, along a specified axis.
data1 = {'id': ['1', '2', '3', '4', '5'],
'Column1': ['A', 'C', 'E', 'G', 'I']}
df1 = pd.DataFrame(data1, columns = ['id', 'Column1'])
data2 = {'id': ['1', '2', '6', '7', '8'],
'Column1': ['K', 'M', 'O', 'Q', 'S'],
'Column2': ['L', 'N', 'P', 'R', 'T']}
df2 = pd.DataFrame(data2, columns = ['id', 'Column1', 'Column2'])
combined_df_rows = pd.concat([df1, df2], ignore_index=True) # Concatenates along the row and adjusts row
# labels automatically with "ignore_index=True"
combined_df_columns = pd.concat([df1, df2], axis=1) # Concatenates along the columns
merge() and join() can do similar operations, but join() is often easier to apply. These are used when DataFrames hold different kinds of information about the same entity, linked by some common feature.
To combine on the common column, use the argument on and set it to the common column name:
data3 = {'id': ['1', '2', '3', '4', '5', '7', '8', '9', '10', '11'],
'Feature3': [12, 13, 14, 15, 16, 17, 15, 12, 13, 23]}
df3 = pd.DataFrame(data3, columns = ['id', 'Feature3'])
merger_df = pd.merge(combined_df_rows, df3, on='id')
Sometimes, the column on which you want to merge has different names. If that is the case, you can specify the arguments left_on and right_on for the left DataFrame and right DataFrame column names, respectively:
data3 = {'identifier': ['1', '2', '3', '4', '5', '7', '8', '9', '10', '11'],
'Feature3': [12, 13, 14, 15, 16, 17, 15, 12, 13, 23]}
df3 = pd.DataFrame(data3, columns = ['identifier', 'Feature3'])
merger_df = pd.merge(combined_df_rows, df3, left_on='id', right_on='identifier')
At the same time, you can also decide what kind of join logics you want to use:
- Full outer join:
how = outer - Inner join:
how = inner - Right join:
how = right - Left join:
how = left
For more information and an easy to visualize guide for these methods, please check this link this link.
Extra exercises¶
Might include usage of other packages, including numpy.
Simple vectorized operations¶
You want to plot the mathematical function
$f(x) = log(-1.3x^2 + 1.4^x + 7x + 50)$
For the numbers in $[0, 20]$. To do this, you need to create a vector xs with lots of numbers between 0 and 20, and a vector ys with $f$ evaluated at every element of xs. A vector is a 1d-ndarray.
To get a hang of vectorized operations, solve the problem without using any loops:
Create a pandas.Series xs with 1000 evenly spaced points between 0 and 20¶
Create a Python function $f$ as seen above¶
Evaluate ys = $f(x)$, i.e. $f$ of every element of xs.¶
What is the mean and standard deviation of ys?¶
How many elements of. ys are below 0? Between 1 and 2, both exclusive?¶
Hint: You can use a comparison operator to get an array of dtype
bool. To get the number of elements that areTrue, you can exploit the fact thatTruebehaves similar to the number 1, andFalsesimilar to the number 0.
What is the minimum and maximum value of ys?¶
Create a series non_negatives, which contain all the values of ys that are nonnegative¶
Extra: Use matplotlib to plot xs vs ys directly from your Series object¶
Species depth matrix¶
Load in the data depths.csv. As you can see in drive preview, there are 11 columns, with columns 2-11 representing a sample from a human git microbiome. Each row represents a genome of a micro-organism, a so-called "operational taxonomic unit at 97% sequence identity" (OTU_97). The first row gives the name of the genome. The values in the matrix represents the relative abundance (or depth) of that micro-organism in that sample, i.e. how much of the micro-organism there is.
Load in the matrix in a pandas.DataFrame¶
url = 'https://raw.githubusercontent.com/Center-for-Health-Data-Science/PythonTsunami/fall2021/data/depths.csv'
depths = pd.read_csv(url)
depths
How many OTUs are there? Show how you figured it out.¶
Find the OTU "OTU_97.41189.0". What is the mean and standard deviations of the depths across the 10 samples of this OTU?¶
How many samples have 0 depth of that OTU? (or rather, below detection limit?)¶
What is the mean and standard deviation if you exclude those samples?¶
Extra: How would you get all the means and std. deviations in one go?¶
We are not interested in OTUs present in fewer than 4 samples. Remove all those OTUs.¶
How many OTUs did you remove?¶
How many OTUs have a depth of > 5 in all 10 samples? (hint: np.all)¶
Filtering and Normalization¶
After discarding all OTUs present in fewer than 4 samples, sort the OTUs, do the following:
Calculate the mean depth across samples for each remaining OTU.
Normalize the remaining OTUs such that each row sum to 0 and have a standard deviation of 1 (so-called z-score normalization)
Print the remaining OTUs to a new file in descending order by their mean depth, with a 12th column giving the mean depth, and columns 1-11 being the normalized depth. Make sure that your file looks like the input file (except with the 12th column)
